crtomo package¶
Subpackages¶
- crtomo.notebook package
- Subpackages
- Submodules
- crtomo.notebook.nb module
crtomo_gui_jupyterdo_we_run_in_ipython()processing_workflow_v1step_inversion_analysisstep_inversion_analysis.apply_next_input()step_inversion_analysis.apply_next_input_from_gui()step_inversion_analysis.can_run()step_inversion_analysis.create_ipywidget_gui()step_inversion_analysis.find_previous_step()step_inversion_analysis.persistency_load()step_inversion_analysis.persistency_store()step_inversion_analysis.set_input_new()step_inversion_analysis.transfer_input_new_to_applied()
step_inversion_settingsstep_inversion_settings.apply_next_input()step_inversion_settings.apply_next_input_from_gui()step_inversion_settings.can_run()step_inversion_settings.create_ipywidget_gui()step_inversion_settings.find_previous_step()step_inversion_settings.persistency_load()step_inversion_settings.persistency_store()step_inversion_settings.set_input_new()step_inversion_settings.transfer_input_new_to_applied()
- Module contents
Submodules¶
crtomo.analytical_solution module¶
Analytical potential distribution over a half-space
For a homogeneous full space, the potential distribution over a point source is given as:
\(\varphi(r) = I \frac{1}{4 \pi \sigma r}\)
- crtomo.analytical_solution.compute_potentials_analytical_hs(grid, configs_raw, rho)[source]¶
Compute the potential superpositions of each current dipole in the configurations, using the provided resistivity
- Parameters:
grid – crt_grid object with loaded FE grid. Used for the electrode positions
configs_raw (numpy.ndarray) – Nx4 array containing N four-point spreads
rho (float) – resistivity of half-space
- Returns:
potentials – List containing N arrays, each of size M (nr of grid nodes)
- Return type:
- crtomo.analytical_solution.compute_voltages(grid, configs_raw, potentials_raw)[source]¶
Given a list of potential distribution and corresponding four-point spreads, compute the voltages
- Parameters:
grid – crt_grid object the grid is used to infer electrode positions
configs_raw (Nx4 array) – containing the measurement configs (1-indexed)
potentials_raw (list with N entries) – corresponding to each measurement, containing the node potentials of each injection dipole.
crtomo.binaries module¶
provide a simple OS-agnostic interface to various binary paths, e.g., for GMSH oder CRTomo.
At the moment all paths are hard-coded, but this interface could be extended to using a configuration file.
At the moment the following binaries have hard-coded default paths:
gmsh
CRTomo
CRMod
CutMcK
Examples
>>> import crtomo.binaries as cBin
gmsh_binary = cBin.get('gmsh')
print(gmsh_binary)
/usr/bin/gmsh'
crtomo.cfg module¶
Representations of CRMod and CRTomo configurations.
Examples
>>> import crtomo.cfg as CRcfg
crmod_cfg = CRcfg.crmod_config()
print(crmod_cfg)
***FILES*** ! mswitch
../grid/elem.dat ! elem
../grid/elec.dat ! elec
../rho/rho.dat ! rho
../config/config.dat ! config
F ! write_pot
../mod/pot/pot.dat ! pot_file
T ! write_volts
../mod/volt.dat ! volt_file
F ! write_sens
../mod/sens/sens.dat ! sens_file
F ! another_dataset
1 ! 2D
F ! fictitious_sink
1660 ! sink_node
F ! boundary_values
boundary.dat ! boundary_file
- class crtomo.cfg.crmod_config(*arg, **kw)[source]¶
Bases:
dictWrite CRMod configuration files (crmod.cfg).
This class is essentially a dict of CRMod configurations with a few extra functions that know how to write a proper crmod.cfg file.
Examples
>>> import crtomo.cfg as cCFG crmod_cfg = cCfg.crmod_config() crmod_cfg.write_to_file('crmod.cfg')
- clear() None. Remove all items from D.¶
- fromkeys(value=None, /)¶
Create a new dictionary with keys from iterable and values set to value.
- get(key, default=None, /)¶
Return the value for key if key is in the dictionary, else default.
- items() a set-like object providing a view on D's items¶
- keys() a set-like object providing a view on D's keys¶
- pop(k[, d]) v, remove specified key and return the corresponding value.¶
If the key is not found, return the default if given; otherwise, raise a KeyError.
- popitem()¶
Remove and return a (key, value) pair as a 2-tuple.
Pairs are returned in LIFO (last-in, first-out) order. Raises KeyError if the dict is empty.
- setdefault(key, default=None, /)¶
Insert key with a value of default if key is not in the dictionary.
Return the value for key if key is in the dictionary, else default.
- update([E, ]**F) None. Update D from dict/iterable E and F.¶
If E is present and has a .keys() method, then does: for k in E: D[k] = E[k] If E is present and lacks a .keys() method, then does: for k, v in E: D[k] = v In either case, this is followed by: for k in F: D[k] = F[k]
- values() an object providing a view on D's values¶
- class crtomo.cfg.crtomo_config(*arg, **kw)[source]¶
Bases:
dictWrite CRTomo configuration files (crtomo.cfg).
This class is essentially a dict of CRTomo configurations with a few extra functions that know how to write a proper crtomo.cfg file.
- clear() None. Remove all items from D.¶
- fromkeys(value=None, /)¶
Create a new dictionary with keys from iterable and values set to value.
- get(key, default=None, /)¶
Return the value for key if key is in the dictionary, else default.
- import_from_file(filename)[source]¶
Import a CRTomo configuration from an existing crtomo.cfg file
- Parameters:
filename (str) – Path to crtomo.cfg file
- items() a set-like object providing a view on D's items¶
- keys() a set-like object providing a view on D's keys¶
- pop(k[, d]) v, remove specified key and return the corresponding value.¶
If the key is not found, return the default if given; otherwise, raise a KeyError.
- popitem()¶
Remove and return a (key, value) pair as a 2-tuple.
Pairs are returned in LIFO (last-in, first-out) order. Raises KeyError if the dict is empty.
- setdefault(key, default=None, /)¶
Insert key with a value of default if key is not in the dictionary.
Return the value for key if key is in the dictionary, else default.
- update([E, ]**F) None. Update D from dict/iterable E and F.¶
If E is present and has a .keys() method, then does: for k in E: D[k] = E[k] If E is present and lacks a .keys() method, then does: for k, v in E: D[k] = v In either case, this is followed by: for k in F: D[k] = F[k]
- values() an object providing a view on D's values¶
crtomo.configManager module¶
Manage measurement configurations and corresponding measurements.
Geometric Factors¶
(Source code, png, hires.png, pdf)
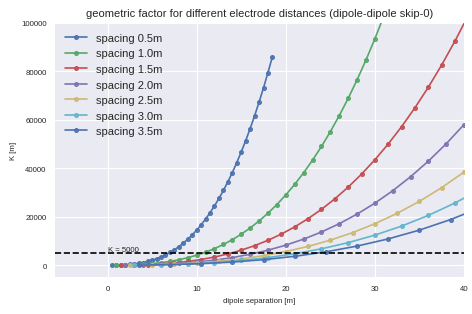
Literature¶
- Noel, Mark and Biwen Xu; Archaeological investigation by electrical
resistivity tomography: a preliminary study. Geophys J Int 1991; 107 (1): 95-102. doi: 10.1111/j.1365-246X.1991.tb01159.x
Stummer, Peter, Hansruedi Maurer, and Alan G. Green. “Experimental Design: Electrical Resistivity Data Sets That Provide Optimum Subsurface Information.” Geophysics 69, no. 1 (January 1, 2004): 120–120. doi:10.1190/1.1649381.
Roy, A., and A. Apparao. “DEPTH OF INVESTIGATION IN DIRECT CURRENT METHODS.” GEOPHYSICS 36, no. 5 (January 1, 1971): 943–59. doi:10.1190/1.1440226.
- class crtomo.configManager.ConfigManager(**kwargs)[source]¶
Bases:
ConfigManager- _crmod_to_abmn(configs)[source]¶
convert crmod-style configurations to a Nx4 array
CRMod-style configurations merge A and B, and M and N, electrode numbers into one large integer each:
\[ \begin{align}\begin{aligned}AB = A \cdot 10^4 + B\\MN = M \cdot 10^4 + N\end{aligned}\end{align} \]- Parameters:
configs (numpy.ndarray) – Nx2 array holding the configurations to convert
Examples
>>> import numpy as np >>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=5) >>> crmod_configs = np.array(( ... (10002, 40003), ... (10010, 30004), ... )) >>> abmn = config._crmod_to_abmn(crmod_configs) >>> print(abmn) [[ 1 2 4 3] [ 1 10 3 4]]
- _get_crmod_abmn()[source]¶
return a Nx2 array with the measurement configurations formatted CRTomo style
- abmn_to_dataframe()[source]¶
Return a pandas.DataFrame containing the measurement configurations
- Returns:
abmn_df – Configurations in a DataFrame (columns: a,b,m,n)
- Return type:
- add_measurements(measurements)[source]¶
Add new measurements to this instance
- Parameters:
measurements (numpy.ndarray) – one or more measurement sets. It must either be 1D or 2D, with the first dimension the number of measurement sets (K), and the second the number of measurements (N): K x N
- Returns:
mid – measurement ID used to extract the measurements later on
- Return type:
Examples
>>> import numpy as np import crtomo.configManager as CRconfig config = CRconfig.ConfigManager(nr_of_electrodes=10) config.gen_dipole_dipole(skipc=0) # generate some random noise random_measurements = np.random.random(config.nr_of_configs) mid = config.add_measurements(random_measurements) # retrieve using mid print(config.measurements[mid])
- add_to_configs(configs)[source]¶
Add one or more measurement configurations to the stored configurations
- Parameters:
configs (list or numpy.ndarray) – list or array of configurations
- Returns:
configs – array holding all configurations of this instance
- Return type:
Kx4 numpy.ndarray
- clear_measurements()[source]¶
Remove all measurements from self.measurements. Reset the measurement counter. All ID are invalidated.
- delete_data_points(indices)[source]¶
Delete data points by index (0-indexed), both in configs and measurements. Deletions will be done in ALL registered measurements to ensure consistency.
- Parameters:
indices (int|iterable) – Indices to delete
- gen_all_current_dipoles()[source]¶
Generate all possible current dipoles for the given number of electrodes (self.nr_electrodes). Duplicates are removed in the process.
After Noel and Xu, 1991, for N electrodes, the number of possible unique configurations is \(N \cdot (N - 1) / 2\). This excludes duplicates in the form of switches current/voltages electrodes, as well as reciprocal measurements.
- Returns:
configs – all possible current dipoles A-B
- Return type:
Nx2 numpy.ndarray
- gen_all_voltages_for_injections(injections_raw)[source]¶
For a given set of current injections AB, generate all possible unique potential measurements.
After Noel and Xu, 1991, for N electrodes, the number of possible voltage dipoles for a given current dipole is \((N - 2)(N - 3) / 2\). This includes normal and reciprocal measurements.
If current dipoles are generated with ConfigManager.gen_all_current_dipoles(), then \(N \cdot (N - 1) / 2\) current dipoles are generated. Thus, this function will produce \((N - 1)(N - 2)(N - 3) / 4\) four-point configurations ABMN, half of which are reciprocals (Noel and Xu, 1991).
All generated measurements are added to the instance.
Use ConfigManager.split_into_normal_and_reciprocal() to split the configurations into normal and reciprocal measurements.
- Parameters:
injections (numpy.ndarray) – Kx2 array holding K current injection dipoles A-B
- Returns:
configs – Nax4 array holding all possible measurement configurations
- Return type:
- gen_configs_permutate(injections_raw, only_same_dipole_length=False, ignore_crossed_dipoles=False, silent=False)[source]¶
Create measurement configurations out of a pool of current injections. Use only the provided dipoles for potential dipole selection. This means that we have always reciprocal measurements.
Remove quadpoles where electrodes are used both as current and voltage dipoles.
- Parameters:
injections_raw (Nx2 array) – current injections
only_same_dipole_length (bool, optional) – if True, only generate permutations for the same dipole length
ignore_crossed_dipoles (bool, optional) – If True, potential dipoles will be ignored that lie between current dipoles, e.g. 1-4 3-5. In this case it is possible to not have full normal-reciprocal coverage.
silent (bool, optional) – if True, do not print information on ignored configs (default: False)
- Returns:
configs – quadrupoles generated out of the current injections
- Return type:
Nx4 array
- gen_dipole_dipole(skipc, skipv=None, stepc=1, stepv=1, nr_voltage_dipoles=10, before_current=False, allow_crossings=False, start_skip=0, N=None)[source]¶
Generate dipole-dipole configurations
- Parameters:
skipc (int) – number of electrode positions that are skipped between electrodes of a given dipole
skipv (int) – steplength between subsequent voltage dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
stepc (int) – steplength between subsequent current dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
stepv (int) – steplength between subsequent voltage dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
nr_voltage_dipoles (int) – the number of voltage dipoles to generate for each current injection dipole
before_current (bool, optional) – if set to True, also generate voltage dipoles in front of current dipoles.
allow_crossings (bool, optional) – Allow potential dipoles to cross current injection dipoles. Note that any quadpole generated by crossing the current electrodes is NOT counted into nr_voltage_dipoles.
start_skip (int, optional) – how many electrode to skip before/after the first/second current electrode.
N (int, optional) – number of electrodes, must be given if not already known by the config instance
Examples
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole(skipc=2) array([[ 1, 4, 5, 8], [ 1, 4, 6, 9], [ 1, 4, 7, 10], [ 2, 5, 6, 9], [ 2, 5, 7, 10], [ 3, 6, 7, 10]])
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole( ... skipc=2, before_current=True, allow_crossings=True) array([[ 1, 4, 2, 5], [ 1, 4, 3, 6], [ 1, 4, 5, 8], [ 1, 4, 6, 9], [ 1, 4, 7, 10], [ 2, 5, 1, 4], [ 2, 5, 3, 6], [ 2, 5, 4, 7], [ 2, 5, 6, 9], [ 2, 5, 7, 10], [ 3, 6, 1, 4], [ 3, 6, 2, 5], [ 3, 6, 4, 7], [ 3, 6, 5, 8], [ 3, 6, 7, 10]])
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole(skipc=1, before_current=True) array([[ 1, 3, 4, 6], [ 1, 3, 5, 7], [ 1, 3, 6, 8], [ 1, 3, 7, 9], [ 1, 3, 8, 10], [ 2, 4, 5, 7], [ 2, 4, 6, 8], [ 2, 4, 7, 9], [ 2, 4, 8, 10], [ 3, 5, 6, 8], [ 3, 5, 7, 9], [ 3, 5, 8, 10], [ 4, 6, 1, 3], [ 4, 6, 7, 9], [ 4, 6, 8, 10], [ 5, 7, 2, 4], [ 5, 7, 1, 3], [ 5, 7, 8, 10]])
- gen_inner_gradients(injections, skips=None)[source]¶
Given a set of current injections, generate voltage dipoles between the current electrodes
Examples
>>> import reda >>> cfg = reda.ConfigManager(nr_of_electrodes=10) >>> cfg.gen_inner_gradients([1, 10], skips=[2, ]) array([[ 1, 10, 2, 5], [ 1, 10, 3, 6], [ 1, 10, 4, 7], [ 1, 10, 5, 8], [ 1, 10, 6, 9]])
- Parameters:
injections (Nx2 numpy.ndarray) – Current injections
skips (None or iterable) – The skips to generate. None will generate all possible skips
- Returns:
configs – The generated configurations
- Return type:
Nx4 numpy.ndarray
- gen_reciprocals(append=False)[source]¶
Generate reciprocal configurations, sort by AB, and optionally append to configurations.
- Parameters:
append (bool) – Append reciprocals to configs (the default is False).
Examples
>>> cfgs = ConfigManager(nr_of_electrodes=5) >>> nor = cfgs.gen_dipole_dipole(skipc=0) >>> rec = cfgs.gen_reciprocals(append=True) >>> print(cfgs.configs) [[1 2 3 4] [1 2 4 5] [2 3 4 5] [3 4 1 2] [4 5 1 2] [4 5 2 3]]
- gen_schlumberger(M, N, a=None)[source]¶
generate one Schlumberger sounding configuration, that is, one set of configurations for one potential dipole MN.
- Parameters:
- Returns:
configs – array holding the configurations
- Return type:
Kx4 numpy.ndarray
Examples
import reda.configs.configManager as CRconfig config = CRconfig.ConfigManager(nr_of_electrodes=40) config.gen_schlumberger(M=20, N=21)
- gen_voltage_dipoles_skip(cinj, skip=0)[source]¶
For given current injections, generate all possible voltage dipoles with skip skip.
- Parameters:
cinj (
numpy.ndarray) – Nx2 array containing the current injection electrodes a,bskip (int, optional) – Skip used for voltage electrodes. Default: 0
- gen_wenner(a)[source]¶
Generate Wenner measurement configurations.
- Parameters:
a (int) – distance (in electrodes) between subsequent electrodes of each four-point configuration.
- Returns:
configs – array holding the configurations
- Return type:
Kx4 numpy.ndarray
- get_unique_current_injections()[source]¶
Return all unique current injection electrode pairs
Examples
>>> from reda.configs.configManager import ConfigManager >>> cfg = ConfigManager() >>> cfg.nr_electrodes >>> cfg.nr_electrodes = 10 >>> cfg.add_to_configs(( ... (1, 2, 3, 4), ... (1, 2, 5, 6), ... (7, 8, 9, 10) ... )) array([[ 1, 2, 3, 4], [ 1, 2, 5, 6], [ 7, 8, 9, 10]]) >>> cfg.get_unique_current_injections() array([[1, 2], [7, 8]])
cfg = ConfigManager() cfg.get_unique_current_injections()
- Returns:
unique_injections – Current injections
- Return type:
Nx2 numpy.ndarray
- load_crmod_config(filename)[source]¶
Load a CRMod configuration file
- Parameters:
filename (string|io.BytesIO) – absolute or relative path to a crmod config.dat file. Can also be a BytesIO object
- load_crmod_data(data_source, is_forward_response=False, try_fix_signs=False)[source]¶
Load CRMod measurement data, either from file or directly from a numpy array.
- Parameters:
data_source (string|numpy.ndarray) – if this is a string, treat it as an input filename. If it is a Nx6 or Nx3 numpy array, use this data directly
is_forward_response (bool, optional) – If True this indicates a volt.dat file created by CRTomo during an inversion run to store forward responses of a given model iteration. In this case the third data column indicates the wdfak parameter, i.e., it indicates if a given data point was excluded from this iteration.
try_fix_signs (bool, optional) – If True, try to fix sign reversals with respect to an already registered configuration set by swappend m and n entries.
- Returns:
cid_mag (int) – Measurement id for magnitude data
cid_pha (int) – Measurement id for phase data
mid_wdfak (int, optional) – Measurement id for wdfak indicators. Only returned if is_forward_response is True
- load_crmod_or_4c_configs(filename)[source]¶
Load configurations either from a CRMod measurement file, or from a four-column file. Assume 1-indexed data (start with electrode 1).
- Parameters:
filename (str) – Path to file that shall be imported
- Return type:
None
- load_crmod_volt(filename)[source]¶
Load a CRMod measurement file (commonly called volt.dat)
- Parameters:
filename (string) – path to input filename
- Returns:
cid_mag (int) – Measurement id for magnitude data
cid_pha (int) – Measurement id for phase data
- load_injections_from_mcf(filename)[source]¶
Load injections from a mcf-file used by the sEIT-systems from FZJ; Injections in the file must be formatted like the following: SE 001 002 SE 002 001 . . .
- Parameters:
filename (string) – absolute or relative path to a mcf-file
- Return type:
Nx2 numpy array with current injections
- property nr_of_configs¶
Return number of configurations
- Returns:
nr_of_configs – number of configurations stored in this instance
- Return type:
- remove_duplicates(configs=None)[source]¶
remove duplicate entries from 4-point configurations. If no configurations are provided, then use self.configs. Unique configurations are only returned if configs is not None.
- Parameters:
configs (Nx4 numpy.ndarray, optional) – remove duplicates from these configurations instead from self.configs.
- Returns:
configs_unique – unique configurations. Only returned if configs is not None
- Return type:
Kx4 numpy.ndarray
- remove_max_dipole_sep(maxsep=10)[source]¶
Remove configurations with dipole separations higher than maxsep.
- Parameters:
maxsep (int) – Maximum separation between both dipoles (the default is 10).
- remove_using_nr_injections(min_nr_of_injections)[source]¶
Remove all measurements with a current injection that is not used a minimum number of times.
This is useful to optimize measurement time for multi-channel systems, such as the IRIS Instruments Syscal Pro. The device can theoretically only facilitate the full number of channels if a corresponding number of voltage measurements is requested for a given current injection. As such, it can be useful to remove measurements with unique current injection dipoles. Note that other factors determine if all channels are actually used. Please refer to the device manual for device-specific information.
- Parameters:
min_nr_of_injections (int) – Minimum number a given current injection should have to keep it
- split_into_normal_and_reciprocal(pad=False, return_indices=False)[source]¶
Split the stored configurations into normal and reciprocal measurements
** Rule 1: the normal configuration contains the smallest electrode number of the four involved electrodes in the current dipole **
- Parameters:
- Returns:
normal (numpy.ndarray) – Nnx4 array. If pad is True, then Nn == N (total number of unique measurements). Otherwise Nn is the number of normal measurements.
reciprocal (numpy.ndarray) – Nrx4 array. If pad is True, then Nr == N (total number of unique measurements). Otherwise Nr is the number of reciprocal measurements.
nor_indices (numpy.ndarray, optional) – Nnx1 array containing the indices of normal measurements. Only returned if return_indices is True.
rec_indices (numpy.ndarray, optional) – Nrx1 array containing the indices of normal measurements. Only returned if return_indices is True.
- to_iris_syscal(filename, spacing=1)[source]¶
Export to IRIS Instrument configuration file
- Parameters:
filename (string) – Path to output filename
- to_pg_scheme(container=None, positions=None)[source]¶
Convert the configuration to a pygimli measurement scheme
- Parameters:
container (reda.containers.ERT.ERT) – an ERT data container (we take the electrode positions from here)
None (positions =)
- Returns:
data
- Return type:
pybert.DataContainerERT
Examples
import numpy as np from reda.configs.configManager import ConfigManager configs = ConfigManager(nr_of_electrodes=48) new_configs = configs.gen_dipole_dipole(skipc=2) x = np.arange(0, 48, 1) z = np.ones(48) * -1 y = np.zeros(48) xyz = np.vstack((x, y, z)).T scheme = configs.to_pg_scheme(positions=xyz) print(scheme)
- property unique_injections¶
- write_crmod_config(filename)[source]¶
Write the configurations to a configuration file in the CRMod format All configurations are merged into one previor to writing to file
- Parameters:
filename (string) – absolute or relative path to output filename (usually config.dat)
- write_crmod_volt(filename, mid)[source]¶
Write the measurements to the output file in the volt.dat file format that can be read by CRTomo.
Data is written in utf-8 encoding.
- Parameters:
filename (string|io.BytesIO) – output filename
mid (int or [int, int]) – measurement ids of magnitude and phase measurements. If only one ID is given, then the phase column is filled with zeros
- write_crmod_volt_with_individual_errors(filename, data_mids, error_mids, norm_mag=1, norm_pha=1)[source]¶
Write the measurements and individual errors to the output file in the volt.dat file format that can be read by CRTomo.
Data is written in utf-8 encoding.
- Parameters:
filename (string|io.BytesIO) – output filename
data_mids ([int, int]) – measurement ids of magnitude and phase measurements
error_mids ([int, int]) – ids of magnitude and phase error estimates
norm_mag (float) – Normalization factor for magnitude errors
norm_pha (float) – Normalization factor for magnitude errors
crtomo.debug module¶
This file contains certain functionality used for testing purposes.
One example is the interface to return grid objects based on CRTomo grids that are distributed with crtomo_tools.
- crtomo.debug.get_grid(key)[source]¶
Return a
crtomo.grid.crt_gridinstance, with a debug grid that is distributed with the crtomo package. Multiple grids are available:key=20 - a 20 electrode grid with 1m spacing, 4 elements between electrodes, rectangular elements.
key=40 - a 40 electrode grid with 1m spacing, 4 elements between electrodes, rectangular elements.
- Parameters:
key (string) – key that identifies the grid
- Returns:
grid – loaded grid object
- Return type:
crtomo.grid.crt_gridinstance
crtomo.eitManager module¶
A digital representation of a sEIT inversion directory.
A sEIT-dir (or sipdir) has the following structure:
- class crtomo.eitManager.eitMan(**kwargs)[source]¶
Bases:
objectManage sEIT data
- __init__(**kwargs)[source]¶
Data/frequency input: Note that only one of the parameters seitdir, crt_data_dir, seit_data will be used during initialization, in the stated priority.
Initialization diagram:
- Parameters:
seitdir (string, optional) – Load frequencies, grid, and data from an existing sEIT directory. Honors the shallow_import parameter.
shallow_import (bool, optional) – In combination with seitdir, only import the last iteration inversion results (faster).
crt_data_dir (string, optional) – if given, then try to load data from this directory. Expect a ‘frequencies.dat’ file and corresponding ‘volt_*.dat’ files.
only_frequency_ids (numpy.ndarray|None) – Load only the frequencies associated with these indices (zero-based). Only works with crt_data_dir.
seit_data (dict) – A dictionary with frequencies as keys, and numpy arrays as items. Each array must have 6 columns: a,b,m,n,magnitude[Ohm],phase[mrad]
frequencies (numpy.ndarray, optional) – frequencies that we work with
grid (crtomo.crt_grid.crt_grid, optional) – A grid instance
elem_file (string, optional) – Path to elem file
elec_file (string, optional) – Path to elec file
decouplings_file (str, optional) – Path to decoupling file: For not will be copied to the exe/ subdir of each tomodir written
crmod_cfg (?, optional)
crtomo_cfg (?, optional)
parman (?, optional)
- _init_frequencies(frequencies, configs_abmn=None)[source]¶
Initialize the tdMan instances associated with each frequency
Note that existing tds will not be deleted/overwritten
- Parameters:
frequencies (Nx1 numpy.ndarray) – Frequencies in ascending order
configs_abmn (None|numpy.ndarray (Mx4)) – Measurement configurations to provide to the generated tdMan instances
- add_homogeneous_model(magnitude, phase=0, frequency=None)[source]¶
Add homogeneous models to one or all tomodirs. Register those as forward models
- add_to_configs(configs)[source]¶
Add configurations to all tomodirs
- Parameters:
configs (
numpy.ndarray) – Nx4 numpy array with abmn configurations
- assign_sip_signatures_using_mask(mask_raw, lookup_table)[source]¶
- Parameters:
lookup_table (dict)
spectrum (sip_response) – SIP spectrum to use for parameterization
- extract_all_spectra(label)[source]¶
Extract all SIP spectra, and return frequencies and a numpy array
- Parameters:
label (str) – the label (data type) to extract. This corresponds to a key in eitMan.assignments. Possible values are rmag, rpha, cre, cim
- extract_points(label, points)[source]¶
Extract data points (i.e., SIP spectra) for one or more points.
- Parameters:
label (str) – the label (data type) to extract. This corresponds to a key in eitMan.assignments. Possible values are rmag, rpha, cre, cim
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
df_all – A dataframe with the extracted data
- Return type:
- get_measurement_responses()[source]¶
Return a dictionary of sip_responses for the modeled SIP spectra
Note that this function does NOT check that each frequency contains the same configurations!
- Returns:
responses – Dictionary with configurations as keys
- Return type:
- load_data_crt_files(data_dict, **kwargs)[source]¶
Load sEIT data from .ctr files (volt.dat files readable by CRTomo, produced by CRMod)
- Parameters:
data_dict (dict) – Data files that are imported. See example down below
Examples
>>> import glob data_files = {} data_files['frequencies'] = 'data/frequencies.dat' files = sorted(glob.glob('data/volt_*.crt')) data_files['crt'] = files
- load_inversion_results(sipdir, shallow_import=True)[source]¶
Given an sEIT inversion directory, load inversion results and store the corresponding parameter ids in self.assignments
Note that all previous data stored in this instance of the eitManager will be overwritten, if required!
- Parameters:
sipdir (string) – path to a CRTomo sip-invdir (i.e., a full sEIT inversion directory)
shallow_import (bool, optional) – If set to True, then only import the last inversion result, nothing else.
- measurements(**kwargs)[source]¶
Return modeled measurements. If not already done, call CRMod for each frequency to actually compute the forward response.
- Returns:
m_all –
dimension: frequency
dimension: config-number
dimension: 2: magnitude and phase (resistivity)
- Return type:
- model(**kwargs)[source]¶
Run the forward modeling for all frequencies.
Use
crtomo.eitManager.eitMan.measurements()to retrieve the resulting synthetic measurement spectra.- Parameters:
**kwargs (dict, optional) – All kwargs are directly provide to the underlying
crtomo.tdManager.tdMan.model()function calls.
- plot_forward_models(maglim=None, phalim=None, **kwargs)[source]¶
Create plots of the forward models
- Returns:
return_dict – Dictionary containing the figure and axes objects of the magnitude plots
- Return type:
- plot_result_spectrum(filename, nr)[source]¶
Plot a given data and inversion response spectrum to file.
WARNING: At this point does not take into account missing configurations for certain frequencies…
- register_forward_model(frequency, mag_model, phase_model)[source]¶
Register a magnitude parameter set and optionally a phase parameter set as a forward model for a given frequency.
- Parameters:
frequency (float) – Frequency. Must match the frequencies in eitMan.tds
mag_model (numpy.ndarray) – The magnitude model (linear scale)
phase_model (numpy.ndarray) – The phase model [mrad]
- save_measurements_to_directory(directory)[source]¶
Store the measurements (either from a previous import, or from forward modeling, in one directory in the CRTomo ‘crt’-style.
Frequencies are stored in a file frequencies.dat.
For each frequency, data is stored in a file volt_%.2i.dat in the CRTomo format.
- save_to_eitdir(directory)[source]¶
Save the eit data into a eit/sip directory structure
- Parameters:
directory (string|path) – output directory
- set_area_to_single_colecole(xmin, xmax, zmin, zmax, ccpars)[source]¶
Parameterize a rectangular area of the forward model using a single-term Cole-Cole response.
crtomo.grid module¶
Manage and plot CRMod/CRTomo grids
Creating grids¶
Grids can be created in various ways:
Regular grids can be created using Griev (which is not supported any more)
Irregular (triangular) grids are created using the command line tool ‘cr_trig_create’
the crt_grid class provides some simple wrapper functions for cr_trig_create that directory load created grids
Notes
After loading an elem.dat file, i.e., the grid, grid information are exposed via various structures:
the header is stored in the dict self.header:
>>> self.header.keys() dict_keys( ['bandwidth', 'element_infos', 'cutmck', 'nr_element_types', 'nr_nodes'] )nodes are stored in self.nodes. Various sortings are available. If in doubt use self.nodes[‘presort’].
elements are stored in self.elements (as node numbers)
Examples
>>> import crtomo.grid as CRTGrid
grid = CRTGrid.crt_grid()
grid.load_grid('elem.dat', 'elec.dat')
>>> # extracting element coordinates in the order of rho.dat file
import crtomo.grid as CRGrid
grid = CRGrid.crt_grid(elem_file='elem.dat', elec_file='elec.dat')
# 1. element (oben links)
grid.nodes['presort'][grid.elements[0], :]
# columns: node number - x position - z position
- class crtomo.grid.crt_grid(elem_file=None, elec_file=None, empty=False)[source]¶
Bases:
objectThe
crt_gridholds and manages one Finite-Element grid.This class provides only basic plotting routines. Advanced plotting routines can be found in
crtomo.plotManager.Examples
>>> import crtomo.grid as CRGrid grid = CRGrid(elem_file='elem.dat', elec_file='elec.dat') print(grid) CRMod/CTRomo grid instance number of elements: 2728 number of nodes: 1440 number of electrodes: 60 grid dimsensions: X: -7.5 37.5 Z: -15.0 0.0
- Wm()[source]¶
Return the smoothing regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.48ff
\[\begin{split}\Psi_m = ||\underline{\underline{W}}_m \underline{m}||^2_2\\ = \sum_{j=1}^M \sum_{i=nb(j)} \alpha_{r_ij} |\frac{m_j - m_i}{\Delta c_{ij}|^2 \Delta b_{ij} \Delta c_{ij}\end{split}\]Note that the anisotropic regularization parameter \(\alpha\) is not implemented yet.
i and j are swapped in the implementation.
See Fig 3.5 in the thesis.
- Wm_mgs(m, beta)[source]¶
Return the MGS regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.50
Note that alpha and f are set 1
- Parameters:
m (numpy.ndarray) – model parameters to compute MGS matrix for
beta (float) – MGS beta parameter
- Returns:
Wm – Sparse MGS matrix
- Return type:
- static _get_area_polygon(points_x, points_z)[source]¶
Return the area of a polygon. The node coordinates must be ordered, but the direction does not matter - we return the abs-value
>>> points_x = [4, 4, 8, 8, -4, -4] >>> points_z = [6, -4, -4, -8, -8, 6] >>> self._get_area_polygon(points_x, points_z) ... 128
- _prepare_grids()[source]¶
depending on the type of grid (rectangular or triangle), prepare grids or triangle lists
- TODO: We want some nice way of not needing to know in the future if we
loaded triangles or quadratic elements.
- _read_elem_elements(fid)[source]¶
Read all FE elements from the file stream. Elements are stored in the self.element_data dict. The keys refer to the element types:
3: Triangular grid (three nodes)
8: Quadrangular grid (four nodes)
11: Mixed boundary element
12: Neumann (no-flow) boundary element
- _read_elem_nodes(fid)[source]¶
Read the nodes from an opened elem.dat file. Correct for CutMcK transformations.
We store three typed of nodes in the dict ‘nodes’:
“raw” : as read from the elem.dat file
“presort” : pre-sorted so we can directly read node numbers from a elec.dat file and use them as indices.
- “sorted”completely sorted as in the original grid (before any
CutMcK)
For completeness, we also store the following keys:
“cutmck_index” : Array containing the indices in “presort” to obtain the “sorted” values: nodes[‘sorted’] = nodes[‘presort’] [nodes[‘cutmck_index’], :]
“rev_cutmck_index” : argsort(cutmck_index)
- analyze_internal_angles(return_plot=False)[source]¶
Analyze the internal angles of the grid. Angles shouldn’t be too small because this can cause problems/uncertainties in the Finite-Element solution of the forward problem. This function prints the min/max values, as well as quantiles, to the command line, and can also produce a histogram plot of the angles.
- Parameters:
return_plot (bool) – if true, return (fig, ax) objects of the histogram plot
- Returns:
fig (matplotlib.figure) – figure object
ax (matplotlib.axes) – axes object
Examples
>>> import crtomo.grid as CRGrid grid = CRGrid.crt_grid() grid.load_elem_file('elem.dat') fig, ax = grid.analyze_internal_angles(Angles) This grid was sorted using CutMcK. The nodes were resorted! Triangular grid found Minimal angle: 22.156368696965796 degrees Maximal angle: 134.99337326279496 degrees Angle percentile 10%: 51.22 degrees Angle percentile 20%: 55.59 degrees Angle percentile 30%: 58.26 degrees Angle percentile 40%: 59.49 degrees Angle percentile 50%: 59.95 degrees Angle percentile 60%: 60.25 degrees Angle percentile 70%: 61.16 degrees Angle percentile 80%: 63.44 degrees Angle percentile 90%: 68.72 degrees generating plot... >>> # save to file with fig.savefig('element_angles.png', dpi=300)
- static create_surface_grid(nr_electrodes=None, spacing=None, electrodes_x=None, electrodes_z=None, depth=None, left=None, right=None, char_lengths=None, lines=None, internal_lines=None, debug=False, workdir=None, force_neumann_only=False)[source]¶
This is a simple wrapper for cr_trig_create to create simple surface grids.
Electrode and boundary positions are rounded to the third digit.
- Parameters:
nr_electrodes (int, optional) – the number of surface electrodes
spacing (float, optional) – the spacing between electrodes, usually in [m], required if nr of electrodes is given
electrodes_x (array, optional) – x-electrode positions can be provided here, e.g., for non-equidistant electrode distances
electrodes_z (array, optional) – z-electrode positions can be provided here, e.g., for non-equidistant electrode distances. Only useful in combination with electrodes_x
depth (float, optional) – the depth of the grid, relative to the minimum z-value. If not given, this is computed as half the maximum distance between electrodes
left (float, optional) – the space allocated left of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
right (float, optional) – the space allocated right of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
char_lengths (float|list of 4 floats, optional) – characteristic lengths, as used by cr_trig_create
lines (list of floats, optional) – at the given depths, add horizontal lines in the grid. Note that all positive values will be multiplied by -1!
internal_lines (list of 4-tuple) – extra lines to add to the grid. Important: These lines must NOT touch the outer edge of the grid (this is not supported by this function, but can be accomplished by manually building the grid)
debug (bool, optional) – default: False. If true, don’t hide the output of cr_trig_create
workdir (string, optional) – if set, use this directory to create the grid. Don’t delete files afterwards.
force_neumann_only (bool, optional) – sometimes we want to use only Neumann boundary conditions. Setting this switch to True will force all boundaries to Neumann. Use only if you know what you are doing! default: False
- Returns:
grid – the generated grid
- Return type:
crtomo.grid.crt_gridinstance
Examples
>>> from crtomo.grid import crt_grid >>> grid = crt_grid.create_surface_grid(40, spacing=0.25, depth=5, ... left=2, right=2, char_lengths=[0.1, 0.5, 0.1, 0.5], ... lines=[0.4, 0.8], debug=False, workdir=None) >>> import pylab as plt >>> fig, ax = plt.subplots() >>> grid.plot_grid_to_ax(ax)
- property element_neighbors¶
- property element_neighbors_old¶
Return a list with element numbers (zero indexed) of neighboring elements. Note that the elements are not sorted. No spacial orientation can be inferred from the order of neighbors.
WARNING: This function is slow due to a nested loop. This would be a good starting point for further optimizations.
In order to speed things up, we could search using the raw data, i.e., with CutMcK enabled sorting, and then restrict the loops to 2x the bandwidth (before - after).
While not being returned, this function also sets the variable self.element_neighbors_edges, in which the common nodes with each neighbor are stored.
- Returns:
neighbors – a list (length equal to nr of elements) with neighboring elements
- Return type:
Examples
- get_electrode_node(electrode)[source]¶
For a given electrode (e.g. from a config.dat file), return the true node number as in self.nodes[‘sorted’]
- get_electrode_positions()[source]¶
Return the electrode positions in an numpy.ndarray
- Returns:
positions – Nx2 array, where N is the number of electrodes. The first column contains x positions, the second z positions.
- Return type:
- get_element_areas()[source]¶
return the areas of the elements
note that this formula is generic, see CRMod code for a simpler one
- Returns:
areas
- Return type:
- get_element_centroids()[source]¶
return the central points of all elements
- Returns:
centroids – Nx2 array x/z coordinates for all (N) elements
- Return type:
- get_element_indices_within_rectangle(xmin, xmax, zmin, zmax)[source]¶
Return the indices of all elements whose center is located within the rectangle defined by the parameters.
The indices can then be used, e.g., to select values from inversion results.
- Parameters:
- Returns:
indices – Array with indices (zero-indexed)
- Return type:
np.array
- get_internal_angles()[source]¶
Compute all internal angles of the grid
- Returns:
NxK array with N the number of elements, and K the number of nodes, filled with the internal angles in degrees
- Return type:
- get_min_max_electrode_distances()[source]¶
Return the minimal and the maximal electrode distance of the grid
- Returns:
amin (float) – minimal electrode distance
amax (float) – maximal electrode distance
- get_minmax()[source]¶
Return min/max x/z coordinates of grid
- Returns:
x ([float, float]) – min, max values of grid dimensions in x direction (sideways)
z ([float, float]) – min, max values of grid dimensions in z direction (downwards)
- static interpolate_grid(ingrid, outgrid, data, method='nearest')[source]¶
Function for interpolating data from one grid to another, using the cell midpoints as datapoint locations. Standard method for interpolating is set to nearest-neighbour.
- Parameters:
ingrid (
crtomo.grid.crt_gridinstance) – CRT grid that matches the input dataoutgrid (
crtomo.grid.crt_gridinstance) – CRT grid to interpolate todata (pandas.dataframe) – input data that matches the input grid (in pandas dataframe format)
method (interpolation method, optional) – Standard interpolation method is nearest-neighbour (‘nearest’). Other possible methods are ‘linear’ and ‘cubic’.
- Returns:
interpolated data – returned dataframe with interpolated data
- Return type:
pandas.dataframe
- load_elem_file(elem_file)[source]¶
Load a CRTomo/CRMod elem.dat mesh file from either a file, or from stringIO
- plot_grid(**kwargs)[source]¶
Plot the mesh
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- Returns:
fig (matplotlib.Figure) – The Figure object
ax (matplotlib.Axes) – The axes object the mesh was plotted to
- plot_grid_to_ax(ax, **kwargs)[source]¶
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- reverse_node_order(element_type)[source]¶
Reverses the order of the nodes of all elements of a given type. This can be used to fix CRTomo/CRMod errors regarding the computation of the determinant during FE system compilation.
Use only if you know what you are doing!
- Parameters:
element_type (int) – Element type to change. Usually 3 or 4 (triangles or quads)
- class crtomo.grid.mesh(elem_file=None, elec_file=None, empty=False)[source]¶
Bases:
crt_grid- Wm()¶
Return the smoothing regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.48ff
\[\begin{split}\Psi_m = ||\underline{\underline{W}}_m \underline{m}||^2_2\\ = \sum_{j=1}^M \sum_{i=nb(j)} \alpha_{r_ij} |\frac{m_j - m_i}{\Delta c_{ij}|^2 \Delta b_{ij} \Delta c_{ij}\end{split}\]Note that the anisotropic regularization parameter \(\alpha\) is not implemented yet.
i and j are swapped in the implementation.
See Fig 3.5 in the thesis.
- Wm_mgs(m, beta)¶
Return the MGS regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.50
Note that alpha and f are set 1
- Parameters:
m (numpy.ndarray) – model parameters to compute MGS matrix for
beta (float) – MGS beta parameter
- Returns:
Wm – Sparse MGS matrix
- Return type:
- __init__(elem_file=None, elec_file=None, empty=False)¶
- static _get_area_polygon(points_x, points_z)¶
Return the area of a polygon. The node coordinates must be ordered, but the direction does not matter - we return the abs-value
>>> points_x = [4, 4, 8, 8, -4, -4] >>> points_z = [6, -4, -4, -8, -8, 6] >>> self._get_area_polygon(points_x, points_z) ... 128
- _prepare_grids()¶
depending on the type of grid (rectangular or triangle), prepare grids or triangle lists
- TODO: We want some nice way of not needing to know in the future if we
loaded triangles or quadratic elements.
- _read_elem_elements(fid)¶
Read all FE elements from the file stream. Elements are stored in the self.element_data dict. The keys refer to the element types:
3: Triangular grid (three nodes)
8: Quadrangular grid (four nodes)
11: Mixed boundary element
12: Neumann (no-flow) boundary element
- _read_elem_header(fid)¶
- _read_elem_neighbors(fid)¶
Read the boundary-element-neighbors from the end of the file
- _read_elem_nodes(fid)¶
Read the nodes from an opened elem.dat file. Correct for CutMcK transformations.
We store three typed of nodes in the dict ‘nodes’:
“raw” : as read from the elem.dat file
“presort” : pre-sorted so we can directly read node numbers from a elec.dat file and use them as indices.
- “sorted”completely sorted as in the original grid (before any
CutMcK)
For completeness, we also store the following keys:
“cutmck_index” : Array containing the indices in “presort” to obtain the “sorted” values: nodes[‘sorted’] = nodes[‘presort’] [nodes[‘cutmck_index’], :]
“rev_cutmck_index” : argsort(cutmck_index)
- _write_elem_header(fid)¶
- _write_elements(fid)¶
- _write_neighbors(fid)¶
- _write_nodes(fid)¶
- add_grid_data(data)¶
Return id
- analyze_internal_angles(return_plot=False)¶
Analyze the internal angles of the grid. Angles shouldn’t be too small because this can cause problems/uncertainties in the Finite-Element solution of the forward problem. This function prints the min/max values, as well as quantiles, to the command line, and can also produce a histogram plot of the angles.
- Parameters:
return_plot (bool) – if true, return (fig, ax) objects of the histogram plot
- Returns:
fig (matplotlib.figure) – figure object
ax (matplotlib.axes) – axes object
Examples
>>> import crtomo.grid as CRGrid grid = CRGrid.crt_grid() grid.load_elem_file('elem.dat') fig, ax = grid.analyze_internal_angles(Angles) This grid was sorted using CutMcK. The nodes were resorted! Triangular grid found Minimal angle: 22.156368696965796 degrees Maximal angle: 134.99337326279496 degrees Angle percentile 10%: 51.22 degrees Angle percentile 20%: 55.59 degrees Angle percentile 30%: 58.26 degrees Angle percentile 40%: 59.49 degrees Angle percentile 50%: 59.95 degrees Angle percentile 60%: 60.25 degrees Angle percentile 70%: 61.16 degrees Angle percentile 80%: 63.44 degrees Angle percentile 90%: 68.72 degrees generating plot... >>> # save to file with fig.savefig('element_angles.png', dpi=300)
- calculate_dimensions()¶
For a regular grid, calculate the element and node dimensions
- static create_surface_grid(nr_electrodes=None, spacing=None, electrodes_x=None, electrodes_z=None, depth=None, left=None, right=None, char_lengths=None, lines=None, internal_lines=None, debug=False, workdir=None, force_neumann_only=False)¶
This is a simple wrapper for cr_trig_create to create simple surface grids.
Electrode and boundary positions are rounded to the third digit.
- Parameters:
nr_electrodes (int, optional) – the number of surface electrodes
spacing (float, optional) – the spacing between electrodes, usually in [m], required if nr of electrodes is given
electrodes_x (array, optional) – x-electrode positions can be provided here, e.g., for non-equidistant electrode distances
electrodes_z (array, optional) – z-electrode positions can be provided here, e.g., for non-equidistant electrode distances. Only useful in combination with electrodes_x
depth (float, optional) – the depth of the grid, relative to the minimum z-value. If not given, this is computed as half the maximum distance between electrodes
left (float, optional) – the space allocated left of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
right (float, optional) – the space allocated right of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
char_lengths (float|list of 4 floats, optional) – characteristic lengths, as used by cr_trig_create
lines (list of floats, optional) – at the given depths, add horizontal lines in the grid. Note that all positive values will be multiplied by -1!
internal_lines (list of 4-tuple) – extra lines to add to the grid. Important: These lines must NOT touch the outer edge of the grid (this is not supported by this function, but can be accomplished by manually building the grid)
debug (bool, optional) – default: False. If true, don’t hide the output of cr_trig_create
workdir (string, optional) – if set, use this directory to create the grid. Don’t delete files afterwards.
force_neumann_only (bool, optional) – sometimes we want to use only Neumann boundary conditions. Setting this switch to True will force all boundaries to Neumann. Use only if you know what you are doing! default: False
- Returns:
grid – the generated grid
- Return type:
crtomo.grid.crt_gridinstance
Examples
>>> from crtomo.grid import crt_grid >>> grid = crt_grid.create_surface_grid(40, spacing=0.25, depth=5, ... left=2, right=2, char_lengths=[0.1, 0.5, 0.1, 0.5], ... lines=[0.4, 0.8], debug=False, workdir=None) >>> import pylab as plt >>> fig, ax = plt.subplots() >>> grid.plot_grid_to_ax(ax)
- determine_path_along_nodes(start_coordinate, end_coordinate)¶
- class element¶
Bases:
object- center()¶
- length_line()¶
- outer_normal_line()¶
- vector_line()¶
Return the vector of the boundary element
- vector_norm_line()¶
- property element_neighbors¶
- property element_neighbors_old¶
Return a list with element numbers (zero indexed) of neighboring elements. Note that the elements are not sorted. No spacial orientation can be inferred from the order of neighbors.
WARNING: This function is slow due to a nested loop. This would be a good starting point for further optimizations.
In order to speed things up, we could search using the raw data, i.e., with CutMcK enabled sorting, and then restrict the loops to 2x the bandwidth (before - after).
While not being returned, this function also sets the variable self.element_neighbors_edges, in which the common nodes with each neighbor are stored.
- Returns:
neighbors – a list (length equal to nr of elements) with neighboring elements
- Return type:
Examples
- find_nearest_node(coords, return_node_coords=True)¶
- get_distance_matrix()¶
- get_electrode_node(electrode)¶
For a given electrode (e.g. from a config.dat file), return the true node number as in self.nodes[‘sorted’]
- get_electrode_positions()¶
Return the electrode positions in an numpy.ndarray
- Returns:
positions – Nx2 array, where N is the number of electrodes. The first column contains x positions, the second z positions.
- Return type:
- get_element_areas()¶
return the areas of the elements
note that this formula is generic, see CRMod code for a simpler one
- Returns:
areas
- Return type:
- get_element_centroids()¶
return the central points of all elements
- Returns:
centroids – Nx2 array x/z coordinates for all (N) elements
- Return type:
- get_element_indices_along_line(p0, p1, N)¶
- get_element_indices_within_rectangle(xmin, xmax, zmin, zmax)¶
Return the indices of all elements whose center is located within the rectangle defined by the parameters.
The indices can then be used, e.g., to select values from inversion results.
- Parameters:
- Returns:
indices – Array with indices (zero-indexed)
- Return type:
np.array
- get_internal_angles()¶
Compute all internal angles of the grid
- Returns:
NxK array with N the number of elements, and K the number of nodes, filled with the internal angles in degrees
- Return type:
- get_min_max_electrode_distances()¶
Return the minimal and the maximal electrode distance of the grid
- Returns:
amin (float) – minimal electrode distance
amax (float) – maximal electrode distance
- get_minmax()¶
Return min/max x/z coordinates of grid
- Returns:
x ([float, float]) – min, max values of grid dimensions in x direction (sideways)
z ([float, float]) – min, max values of grid dimensions in z direction (downwards)
- get_node_tree()¶
- get_polygon_from_file(filename)¶
- static interpolate_grid(ingrid, outgrid, data, method='nearest')¶
Function for interpolating data from one grid to another, using the cell midpoints as datapoint locations. Standard method for interpolating is set to nearest-neighbour.
- Parameters:
ingrid (
crtomo.grid.crt_gridinstance) – CRT grid that matches the input dataoutgrid (
crtomo.grid.crt_gridinstance) – CRT grid to interpolate todata (pandas.dataframe) – input data that matches the input grid (in pandas dataframe format)
method (interpolation method, optional) – Standard interpolation method is nearest-neighbour (‘nearest’). Other possible methods are ‘linear’ and ‘cubic’.
- Returns:
interpolated data – returned dataframe with interpolated data
- Return type:
pandas.dataframe
- load_elec_file(elec_file)¶
- load_elem_file(elem_file)¶
Load a CRTomo/CRMod elem.dat mesh file from either a file, or from stringIO
- load_grid(elem_file, elec_file)¶
Load elem.dat and elec.dat
- plot_grid(**kwargs)¶
Plot the mesh
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- Returns:
fig (matplotlib.Figure) – The Figure object
ax (matplotlib.Axes) – The axes object the mesh was plotted to
- plot_grid_to_ax(ax, **kwargs)¶
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- reverse_node_order(element_type)¶
Reverses the order of the nodes of all elements of a given type. This can be used to fix CRTomo/CRMod errors regarding the computation of the determinant during FE system compilation.
Use only if you know what you are doing!
- Parameters:
element_type (int) – Element type to change. Usually 3 or 4 (triangles or quads)
- save_elec_file(filename)¶
- save_elem_elec_files(file_elem='elem.dat', file_elec='elec.dat')¶
- save_elem_file(output)¶
Save elem.dat to file. The grid is saved as read in, i.e., with or without applied cutmck. If you want to change node coordinates, use self.nodes[‘raw’]
- Parameters:
filename (string) – output filename
- test_plot()¶
crtomo.interface module¶
This is meant as a simple interface to CRMod, i.e. the forward operator and its Jacobian. Nothing more.
It can be used to experiment with alternative inversion strategies outside of the old FORTRAN code, but bear in mind that the CRMod binary is used to compute everything. This is slow and involves writing things to disc. If possible use another, better interfaced forward code such as pygimli.
The plan is to support both resistivity-only and complex (magnitude/phase) versions.
- class crtomo.interface.crmod_interface(grid, configs, tempdir=None, crmod_cfg=None)[source]¶
Bases:
object- J(log_sigma, silent=True)[source]¶
Return the sensitivity matrix
- Parameters:
log_sigma (numpy.ndarray) – log_e conductivities
- J_complex_Y_sigma(sigma_model_linear)[source]¶
Compute the complex sensitivity matrix for \(\frac{\del V}{\del \sigma}\).
- Parameters:
sigma_model_linear (numpy.ndarray) – complex linear conductivities
- Returns:
J_complex_lin – Jacobian
- Return type:
NxM numpy.ndarray
- J_complex_logY_sigma(sigma)[source]¶
Return the sensitivity matrix
At this point works only with resistivities.
- Parameters:
sigma (numpy.ndarray) – log_e conductivities
- __init__(grid, configs, tempdir=None, crmod_cfg=None)[source]¶
Initialize the CRMod interface
- Parameters:
grid (crtomo.grid.crt_grid) – FE grid to work with
configs (Nx4 numpy.ndarray) – Measurement configurations to work with (ABMN)
tempdir (string, optional) – If set, use this directory for (temporary) file output. For example using the RAM-disc (/dev/shm) can potentially speed things up a lot.
- _get_tdm(m)[source]¶
For a given model of resistivity magnitudes and phases, return a tdMan instance
- Parameters:
m (Nx1|Nx2 ndarray) – N Model parameters, first column linear resistivity magnitudes [ohm m], second column resistivity phase values [mrad]
- Returns:
tdm – td manager
- Return type:
- forward_complex(log_sigma, silent=True)[source]¶
Compute a model response, i.e. complex impedances
- Parameters:
log_sigma (1xN or 2xN numpy.ndarray) – Model parameters. First column: \(log_e(\sigma)\), second column: \(\phi_{cond} [mrad]\). If first dimension is of length one, assume phase values to be zero.
silent (bool (True)) – If True, suppress output of CRMod
- Returns:
measurements – Return log_e Y values of computed forward response
- Return type:
Nx2 numpy.ndarray
- fwd_complex_R_rho(model_rcomplex, silent=False)[source]¶
Compute the forward response.
- Parameters:
model_rcomplex (size M numpy.ndarray) – Complex forward model, complex resistivities (linear)
- Returns:
response – Model response, complex impedances
- Return type:
- fwd_complex_R_sigma(model_ccomplex, silent=False)[source]¶
Compute the model response from linear complex conductivities
- fwd_complex_logY_sigma(sigma)[source]¶
Compute a model response from linear complex conductivities
- Parameters:
sigma (1xN or 2xN numpy.ndarray) – Model parameters as sigma and sigma phase, N the number of cells. If first dimension is of length one, assume phase values to be zero
- Returns:
measurements – Return log_e sigma values of computed forward response (i.e., first column: log(sigma), second column: sigma phase
- Return type:
Nx2 numpy nd array
crtomo.mpl module¶
This file set ups matplotlib plot functions for the whole package.
Import all necessary Matplotlib modules and set default options To use this module, call the setup() function.
Examples
>>> import crtomo.mpl
>>> crtomo.mpl.setup()
- crtomo.mpl.mpl_get_cb_bound_below_plot(ax)[source]¶
Return the coordinates for a colorbar axes below the provided axes object. Take into account the changes of the axes due to aspect ratio settings.
Important: Use only AFTER fig.subplots_adjust(…)
- crtomo.mpl.mpl_get_cb_bound_next_to_plot(ax)[source]¶
Return the coordinates for a colorbar axes next to the provided axes object. Take into account the changes of the axes due to aspect ratio settings.
Parts of this code are taken from the transforms.py file from matplotlib
Important: Use only AFTER fig.subplots_adjust(…)
Examples
>>> cb_pos = mpl_get_cb_bound_next_to_plot(fig.axes[15]) ax1 = fig.add_axes(cb_pos, frame_on=True) cmap = mpl.cm.jet_r norm = mpl.colors.Normalize(vmin=float(23), vmax=float(33)) # cmap = pm.cmap # norm = pm.norm cb1 = mpl.colorbar.ColorbarBase( ax1, cmap=cmap, norm=norm, orientation='vertical' ) cb1.locator = mpl.ticker.FixedLocator([23, 28, 33]) cb1.update_ticks() cb1.ax.artists.remove(cb1.outline) # remove framei
crtomo.mpl_setup module¶
This file set ups matplotlib plot functions for the whole package.
Import all necessary Matplotlib modules and set default options To use this module, import * from it:
Examples
>>> from crtomo.mpl_setup import *
- crtomo.mpl_setup.mpl_get_cb_bound_below_plot(ax)[source]¶
Return the coordinates for a colorbar axes below the provided axes object. Take into account the changes of the axes due to aspect ratio settings.
Important: Use only AFTER fig.subplots_adjust(…)
- crtomo.mpl_setup.mpl_get_cb_bound_next_to_plot(ax)[source]¶
Return the coordinates for a colorbar axes next to the provided axes object. Take into account the changes of the axes due to aspect ratio settings.
Parts of this code are taken from the transforms.py file from matplotlib
Important: Use only AFTER fig.subplots_adjust(…)
Examples
>>> cb_pos = mpl_get_cb_bound_next_to_plot(fig.axes[15]) ax1 = fig.add_axes(cb_pos, frame_on=True) cmap = mpl.cm.jet_r norm = mpl.colors.Normalize(vmin=float(23), vmax=float(33)) # cmap = pm.cmap # norm = pm.norm cb1 = mpl.colorbar.ColorbarBase( ax1, cmap=cmap, norm=norm, orientation='vertical' ) cb1.locator = mpl.ticker.FixedLocator([23, 28, 33]) cb1.update_ticks() cb1.ax.artists.remove(cb1.outline) # remove framei
crtomo.nodeManager module¶
Manage node value sets for a given grid. Node data assigns a numeric value to each node of a grid. This could, for example, be potential distributions or sensitivities for single files.
- class crtomo.nodeManager.NodeMan(grid_obj)[source]¶
Bases:
objectmanage one or more node value sets for a given
crtomo.grid.crt_gridobject- __init__(grid_obj)[source]¶
- Parameters:
grid_obj (crtomo.grid.crt_grid) – The FE grid that the parameter sets refer to
- add_data(data)[source]¶
Add data to the node value sets
- Parameters:
data (numpy.ndarray) – one or more node value sets. It must either be 1D or 2D, with the first dimension the number of parameter sets (K), and the second the number of elements (Z): K x Z
Examples
>>> # suppose that grid is a fully initialized grid oject with 50 nodes nodeman = NodeMan(grid) # one_data_set = np.ones(50) cid = nodeman.add_data(one_data_set) print(nodeman.parsets[cid]) two_data_sets = np.ones((2, 50)) cids = nodeman.add_data(two_data_sets) print(cids) [0, ] [1, 2]
crtomo.parManager module¶
Manage parameter sets for a given grid. A parameter set is a set of values that correspond to the elements of the grid. Usually this is a resistivity or phase distribution, but can also be a cumulated sensitivity distribution.
- class crtomo.parManager.ParMan(grid_obj)[source]¶
Bases:
objectmanage one or more parameter sets for a given
crtomo.grid.crt_gridobject- __init__(grid_obj)[source]¶
- Parameters:
grid_obj (crtomo.grid.crt_grid) – The FE grid that the parameter sets refer to
- _clean_pid(pid)[source]¶
if pid is a number, don’t do anything. If pid is a list with one entry, strip the list and return the number. If pid contains more than one entries, do nothing.
- add_2d_cos_anomaly_line(pid, p0, anomaly_width, anomaly_height, peak_value, area='only_one_y', only_one_y_line=True, only_one_x_line=False, whole_mesh=False)[source]¶
Add one line of cos(x)cos(y) anomalies to a given parameter set. The wavelength refers to half a period, with the maximum of the anomaly centered on p0=[x0, z0].
- Parameters:
whole_mesh (bool, optional) – If True, then fill the whole mesh with a cos(x)cos(y) pattern.
- add_checkerboard_pattern(pid, p0, anomaly_width, anomaly_height, peak_value)[source]¶
Note that if p0 is larger than a few anomaly sizes its possible that the checkerboard pattern will only be applied to parts of the mesh!
- add_data(data, metadata=None)[source]¶
Add data to the parameter set
- Parameters:
data (numpy.ndarray) – one or more parameter sets. It must either be 1D or 2D, with the first dimension the number of parameter sets (K), and the second the number of elements (N): K x N
metadata (object, optional) – the provided object will be stored in in the metadata dict and can be received with the ID that is returned. If multiple (K) datasets are added at ones, provide a list of objects with len K.
- Returns:
ID which can be used to access the parameter set
- Return type:
int, ID
Examples
>>> # suppose that grid is a fully initialized grid oject with 100 # elements parman = ParMan(grid) # one_data_set = np.ones(100) cid = parman.add_data(one_data_set) print(parman.parsets[cid]) two_data_sets = np.ones((2, 100)) cids = parman.add_data(two_data_sets) print(cids) [0, ] [1, 2]
- add_empty_dataset(value=1)[source]¶
Create an empty data set. Empty means: all elements have the same value.
- Parameters:
value (float, optional) – which value to assign all element parameters. Default is one.
- add_gaussian_anomaly_to_parset(pid, center, width, max_value)[source]¶
- center_of_mass_value(pid, mode='log10')[source]¶
Compute the center of mass value of a given parameter set.
- create_parset_with_gaussian_anomaly(center, width, max_value, background)[source]¶
- Parameters:
- Returns:
pid – ID of newly created parameter set
- Return type:
- extract_along_line(pid, xy0, xy1, N=10)[source]¶
Extract parameter values along a given line.
- Parameters:
- Returns:
values – data values for extracted data points. First column: x-coordinates, second column: z-coordinates, third column: extracted values
- Return type:
numpy.ndarray (n x 3)
- extract_points(pid, points)[source]¶
Extract values at certain points in the grid from a given parameter set. Cells are selected by interpolating the centroids of the cells towards the line using a “nearest” scheme.
Note that data is only returned for the points provided. If you want to extract multiple data points along a line, defined by start and end point, use the extract_along_line function.
- Parameters:
pid (int) – The parameter id to extract values from
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
values – data values for extracted data points
- Return type:
numpy.ndarray (n x 1)
- extract_polygon_area(pid, polygon_points)[source]¶
Extract all data points whose element centroid lies within the given polygon.
- Parameters:
- Returns:
elements_in_polygon_array (numpy.ndarray) – list with elements numbers (zero-indexed, starting from 0) which lie within the polygon
values (list) – the data values corresponding to the elements in elements_in_polygon
- load_from_rho_file(filename)[source]¶
Convenience function that loads two parameter sets from a rho.dat file, as used by CRMod for forward resistivity/phase models.
- Parameters:
filename (string, file path) – filename of rho.dat file
- Returns:
cid_mag (int) – ID of magnitude parameter set
cid_phase (int) – ID of phase parameter set
- load_from_sens_file(filename)[source]¶
Load real and imaginary parts from a sens.dat file generated by CRMod
- Parameters:
filename (string) – filename of sensitivity file
- Returns:
nid_re (int) – ID of real part of sensitivities
nid_im (int) – ID of imaginary part of sensitivities
- load_inv_result(filename, columns=2, is_log10=False)[source]¶
Load one parameter set from a rho*.mag or rho*.pha file produced by CRTomo.
- Parameters:
- Returns:
pid – ID(s) of parameter set
- Return type:
- load_model_from_file(filename, columns=0)[source]¶
Load one parameter set from a file which contains one value per line
No row is skipped.
- modify_area(pid, xmin, xmax, zmin, zmax, value)[source]¶
Modify the given dataset in the rectangular area given by the parameters and assign all parameters inside this area the given value.
Partially contained elements are treated as INSIDE the area, i.e., they are assigned new values.
- Parameters:
pid (int) – id of the parameter set to modify
xmin (float) – smallest x value of the area to modify
xmax (float) – largest x value of the area to modify
zmin (float) – smallest z value of the area to modify
zmin – largest z value of the area to modify
value (float) – this value is assigned to all parameters of the area
Examples
>>> import crtomo.tdManager as CRtdm tdman = CRtdm.tdMan( elem_file='GRID/elem.dat', elec_file='GRID/elec.dat', ) pid = tdman.parman.add_empty_dataset(value=1) tdman.parman.modify_area( pid, xmin=0, xmax=2, zmin=-2, zmin=-0.5, value=2, ) fig, ax = tdman.plot.plot_elements_to_ax(pid) fig.savefig('out.png')
- modify_polygon(pid, polygon, value)[source]¶
Modify parts of a parameter set by modifying all elements within a provided
shapely.geometry.Polygoninstance. Hereby, an element is modified if its center lies within the polygon.- Parameters:
Examples
>>> import shapely.geometry polygon = shapely.geometry.Polygon(( (2, 0), (4, -1), (2, -1) )) tdman.parman.modify_polygon_centroids(pid, polygon, 3)
- modify_polygon_old(pid, polygon, value)[source]¶
Modify parts of a parameter set by setting all parameters located in, or touching, the provided
shapely.geometry.Polygoninstance.WARNING: This implementation often leads to ragged borders in the selected polygons. Use the new modify_polygon function!
- Parameters:
Examples
>>> import shapely.geometry polygon = shapely.geometry.Polygon(( (2, 0), (4, -1), (2, -1) )) tdman.parman.modify_polygon(pid, polygon, 3)
- reset()[source]¶
Resets the ParMan instance. This process deletes all data and metadata, and resets the index variable
crtomo.plotManager module¶
Manage node and element plots
- crtomo.plotManager.converter_change_sign(data)[source]¶
Reverse the sign of the data. Useful for plotting phase values
- crtomo.plotManager.converter_pm_log10(data, verbose_return=False)[source]¶
Convert the given data to:
log10(subdata) for subdata > 0 log10(-subdata’) for subdata’ < 0 0 for subdata’’ == 0
- Parameters:
data (array) – input data
verbose_return (bool) – if True, then also return the indices for cells larger/smaller than zero
- Returns:
array_converted – converted data
- Return type:
array
- class crtomo.plotManager.plotManager(**kwargs)[source]¶
Bases:
objectThe
plotManagerproduces plots for a given grid. It usescrtomo.grid.crt_gridto manage the grid, andcrtomo.parManagerto manage element parameter values.crtomo.nodeManageris used to manage node data.- __init__(**kwargs)[source]¶
initialize the plot manager. Multiple combinations are possible for the kwargs.
- Parameters:
grid (crtomo.grid.crt_grid, optional) – a fully initialized grid object. If not provided, elem and elec files must be provided!
elem_file (file path) – file path to an elem.dat FE grid file. If no grid is provided, a new crt_grid oject is initialized
elec_file (file path) – file path to an elec.dat FE electrode file
nm (crtomo.nodeManager.nodeMan instance, optional) – node manager for node data. If none is provided, an empty one is initialized
pm (crtomo.parManager.parMan instance, optional) – parameter manager for element data. If none is provided, an empty one is initialized
- plot_elements_to_ax(cid, ax=None, **kwargs)[source]¶
Plot element data (parameter sets).
If the parameter ax is not set, then a new figure will be created with a corresponding axes.
- Parameters:
cid (int or
numpy.ndarray) – if cid is an int, then treat it as the id of the parameter set stored in self.parman. Otherwise, expect it to be the data to plot. At the moment no checks are made that the data fits the grid.ax (matplotlib.Axes, optional) – plot to this axes object, if provided
cid_alpha (int, optional) – if given, use the corresponding dataset in self.parman as the alpha channel. No checks are made if all values of this data set lie between 0 and 1 (0 being fully transparent, and 1 being opaque).
xmin (float, optional) – minimal x limit to plot
xmax (float, optional) – maximal x limit to plot
zmin (float, optional) – minimal z limit to plot
zmax (float, optional) – maximial z limit to plot
converter (function, optional) – if given, then use this function to convert the data into another representation. The given function must work with a numpy array. Default: None
norm (norm object, optional) – the norm object for matplotlib plotting can be provided here
plot_colorbar (bool, optional) – if true, plot a colorbar next to the plot
cmap_name (string, optional) – name of the colorbar to use. Default is “viridis”. To reverse colors, use the _r version “viridis_r”
cbposition –
?
cblabel (string, optional) – colorbar label
cbsegments (int, optional) –
?
cbnrticks (int, optional) –
?
over (color, optional) – color to use for values above the current cb-limit. Default: ?
under – color to use for values below the current cb-limit. Default: ?
bad – color to use for nan-values. Default: ?
title (string, optional) – plot title string
xlabel (string, optional) – Set xlabel of the resulting plot
ylabel (string, optional) – Set ylabel of the resulting plot
no_elecs (bool, optional) – If True, plot no electrodes
rasterize (bool, optional) – if True, rasterize the plot. Default: False
aspect ('auto'|'equal', optional default: 'equal') – Aspect of the plot region
cb_pad (float, optional) – Padding of colorbar. Defaults to 0.4
- Returns:
fig
ax
cnorm
cmap
cb (colorbar instance, optional) – only of plot_colorbar is True
scalarMap – use to create custom colorbars
- plot_nodes_contour_to_ax(ax, nid, **kwargs)[source]¶
Plot node data to an axes object
- Parameters:
ax (axes object, optional) – axes to plot to. If not provided, generate a new figure and axes
nid (int) – node id pointing to the respective data set
cmap (string, optional) – color map to use. Default: jet
cbmin (float, optional) – Minimum colorbar value
cbmax (float, optional) – Maximum colorbar value
plot_colorbar (bool, optional) – if true, plot a colorbar next to the plot
use_aspect (bool, optional) – plot grid in correct aspect ratio. Default: True
fill_contours (bool, optional (true)) – If True, the fill the contours (contourf)
crtomo.status module¶
- crtomo.status.is_sipdir(directory)[source]¶
Simple check if the supplied directory is a SIP directory.
- Parameters:
directory (string) – Check if the supplied path is a valid SIP directory
- Returns:
is_sipdir – True if the supplied directory is a SIP directory
- Return type:
- crtomo.status.is_tomodir(directory)[source]¶
Check if the supplied directory is a tomodir
- Parameters:
directory (string) – Check if the supplied path is a valid tomodir
- Returns:
is_tomodir – True if the supplied directory is a tomodir directory
- Return type:
- crtomo.status.sipdir_is_finished(sipdir)[source]¶
Return the state of modeling and inversion for a given SIP dir. The result does not take into account sensitivities or potentials, as optionally generated by CRMod.
- Parameters:
sipdir (string) – Directory to check
- Returns:
crmod_is_finished (bool) – True if all tomodirs of this SIP directory contain finished modeling results.
crtomo_is_finished (bool) – True if all tomodirs of this SIP directory contain finished inversion results.
- crtomo.status.td_is_finished(tomodir)[source]¶
Return the state of modeling and inversion for a given tomodir. The result does not take into account sensitivities or potentials, as optionally generated by CRMod.
- Parameters:
tomodir (string) – Directory to check
- Returns:
crmod_is_finished (bool) – True if a successful CRMod result is contained in the tomodir directory.
crtomo_is_finished (bool) – True if a successful CRTomo inversion results is contained in the tomodir directory.
crtomo.tdManager module¶
A digital representation of a tomodir, i.e., a single-frequency inversion.
** Modelling **
save to one large binary file
I suggest to use pytables (tables) for that.
need to figure out how to store the individual modules in there, and retrieve them later.
save-to-tomodir
including modeling results
on-demand generation of modeling results
provide easy plotting of pseudo sections for voltages:
magnitude (linear/log)
phase (liner)
rho (analytic/numerical K factor)
sigma’
sigma’’
plots should be generated for dipole-dipole only, and all additional configurations that we can find reliable z-estimators for.
Provide corresponding filter functions, or use reda for that (do we want to depend on reda? I think it’s ok to do this).
In case only a subset of data is plotted, note the number of missing values in the plot!
We could also provide means to generate the (x/z) coordinates by means of averaging over sensitivity distributions (see old script for that).
provide functionality to check voltages against sensitivity sums
plot sensitivities
perhaps provide different transformation functions, for example the one from Johannes Kenkel
plot potential distributions
also plot current lines via streamlines?
can we generate current densities? not sure how to mix element data (sigma) and potential data (nodes) in j = sigma E
- class crtomo.tdManager.noise_model(seed, mag_rel=0, mag_abs=0, pha_a1=0, pha_b1=0, pha_rel=0, pha_abs=0)[source]¶
Bases:
objectnoise model as contained in the crt.noisemod file
Notes
line: 1 # Ensemble seed
line: 0.300 # Relative error resistance A (noise) [%] dR=AR+B
line: 0.100E-01 # Absolute errior resistance B (noise) [Ohm m]
line: 0.00 # Phase error parameter A1 [mRad/Ohm/m] dp=A1*R^B1+A2*p+p0
line: 0.00 # Phase error parameter B1 (noise) []
line: 0.00 # Relative phase error A2 (noise) [%]
line: 0.00 # Absolute phase error p0 (noise) [mRad]
- class crtomo.tdManager.tdMan(**kwargs)[source]¶
Bases:
objectManage tomodirs
- __init__(**kwargs)[source]¶
The following initialization permutations are possible:
initialize empty
- initialize from an existing tomodir
tomodir: [tomodir path]
- supply one or more of the building blocks of a tomodir
grid: crtomo.grid.crt_grid instance
crmod_cfg: crtomo.cfg.crmod_cfg instance
crtomo_cfg: crmod.cfg.crtomo_cfg instance
http://www2.geo.uni-bonn.de/~mweigand/dashboard/content/crtomo_doc/crtomo/files.html#inv
- Keyword Arguments:
grid (crtomo.grid.crt_grid) – A fully initialized grid object
tempdir (string|None, optional) – if set, use this directory to create all temporary directories in. For example, settings this to /dev/shm can result in faster generation of large Jacobians
volt_file (None|str|numpy.ndarray) – if this is None, assume we didn’t get any measurement data. If this is a str, assume it to be the filename to a CRTomo volt.dat file. If it is a numpy array, assume 6 columns: a,b,m,n,rmag,rpha
volt_data (synonym for volt_file parameter) – see description for volt_file
- _initialize_components(kwargs)[source]¶
initialize the various components using the supplied **kwargs
- Parameters:
kwargs (dict) – kwargs dict as received by __init__()
- _invert(tempdir, catch_output=True, **kwargs)[source]¶
Internal function than runs an inversion using CRTomo.
- Parameters:
- Returns:
success (bool) – False if an error was detected
error_msg (str) – Error message. None if not error was encountered.
output (str) – Output of the actual CRTomo call
- static _read_eps_ctr(tomodir)[source]¶
Parse a CRTomo eps.ctr file.
TODO: change parameters to only provide eps.ctr file
- Parameters:
tomodir (string) – Path to directory path
- _read_inv_ctr(tomodir)[source]¶
Read in selected results of the inv.ctr file
- Parameters:
tomodir (string) – directory path to a tomodir
- Returns:
inv_ctr – structure containing inv.ctr data
- Return type:
?
- _read_inversion_fwd_responses(tomodir)[source]¶
Import the forward responses for all iterations of a given inversion
- Parameters:
tomodir (str) – Path to tomodir
- _read_inversion_results(tomodir)[source]¶
Import resistivity magnitude/phase and real/imaginary part of conductivity for all iterations
- Parameters:
tomodir (str) – Path to tomodir
- _read_l1_coverage(tomodir)[source]¶
Read in the L1 data-weighted coverage (or cumulated sensitivity) of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_l2_coverage(tomodir)[source]¶
Read in the L2 data-weighted coverage (or cumulated sensitivity) of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_modeling_results(mod_directory, silent=False)[source]¶
Read modeling results from a given mod/ directory. Possible values to read in are:
voltages
potentials
sensitivities
- _read_resm_m(tomodir)[source]¶
Read in the diagonal entries of the resolution matrix of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_sensitivities(sens_dir)[source]¶
import sensitivities from a directory
Note
check that signs are correct in case CRMod switches potential electrodes
- add_homogeneous_model(magnitude, phase=0)[source]¶
Add a homogeneous resistivity model to the tomodir. This is useful for synthetic measurements.
- Parameters:
- Returns:
pid_mag (int) – ID value of the parameter set of the magnitude model
pid_pha (int) – ID value of the parameter set of the phase model
Note that the parameter sets are automatically registered as the
forward models for magnitude and phase values.
- check_measurements_against_sensitivities(magnitude, phase=0, return_plot=False)[source]¶
Check for all configurations if the sensitivities add up to a given homogeneous model
- Parameters:
- Returns:
results (Nx6 numpy.ndarray) – Results of the analysis.
magnitude measurement [Ohm]
sum of sensitivities [Volt]
relative deviation of sensitivity-sum from measurement [in percent]
fig (matplotlib.figure, optional) – figure object. Only returned of return_plot=True
axes (list) – list of axes corresponding to the figure
Examples
>>> #!/usr/bin/python import crtomo.tdManager as CRtdMan tdm = CRtdMan.tdMan( elem_file='grid/elem.dat', elec_file='grid/elec.dat', config_file='config/config.dat', ) results, fig, axes = tdm.check_measurements_against_sensitivities( magnitude=100, phase=-10, return_plot=True ) fig.savefig('sensitivity_comparison.png', dpi=300)
- copy()[source]¶
Provide a copy of yourself. Do not copy modeling or inversion results, but copy everything that can be used for modeling or inversion
- extract_along_line(pid, xy0, xy1, N=10)¶
Extract parameter values along a given line.
- Parameters:
- Returns:
values – data values for extracted data points. First column: x-coordinates, second column: z-coordinates, third column: extracted values
- Return type:
numpy.ndarray (n x 3)
- extract_points(pid, points)¶
Extract values at certain points in the grid from a given parameter set. Cells are selected by interpolating the centroids of the cells towards the line using a “nearest” scheme.
Note that data is only returned for the points provided. If you want to extract multiple data points along a line, defined by start and end point, use the extract_along_line function.
- Parameters:
pid (int) – The parameter id to extract values from
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
values – data values for extracted data points
- Return type:
numpy.ndarray (n x 1)
- get_fwd_reda_container()[source]¶
Return a REDA container, either reda.ERT, or reda.CR, with modeled data
- Returns:
container
- Return type:
reda.ERT|reda.CR|None
- get_potential(config_nr)[source]¶
Return potential data for a given measurement configuration.
- Parameters:
config_nr (int) – Number of the configurations. Starts at 0
- Returns:
pot_data – First array: magnitude potentials, second array: phase potentials
- Return type:
list with two numpy.ndarrays
- get_sensitivity(config_nr)[source]¶
return a sensitivity, as well as corresponding metadata, for a given measurement configuration. Indices start at zero.
- inv_get_last_pid(parameter)[source]¶
Return the pid of the parameter set corresponding to the final inversion results of a given parameter. Return None if the parameter type does not exist, or no inversion result was registered.
- inv_last_cim_parset()[source]¶
- Return the imaginary part of the complex resistivity of the last
inversion iteration.None if no inversion data exists.
- Returns:
inv_last_cim
- Return type:
numpy.ndarray|None
- inv_last_cre_parset()[source]¶
- Return the real part of the complex resistivity of the last
inversion iteration. None if no inversion data exists.
- Returns:
inv_last_cre
- Return type:
numpy.ndarray|None
- inv_last_rmag_parset()[source]¶
Return the resistivity magnitude of the last iteration. None if no inversion data exists.
Example
>>> import crtomo ... tdm = crtomo.tdMan('tomodir/') ... tdm.inv_last_rmag_parset()
- Returns:
inv_last_rmag
- Return type:
numpy.ndarray|None
- inv_last_rpha_parset()[source]¶
- Return the phase magnitude of the last inversion iteration.
None if no inversion data exists.
- Returns:
inv_last_rpha
- Return type:
numpy.ndarray|None
- invert(output_directory=None, catch_output=True, **kwargs)[source]¶
Invert this instance, and import the result files
No directories/files will be overwritten. Raise an IOError if the output directory exists.
- Parameters:
output_directory (string, optional) – use this directory as output directory for the generated tomodir. If None, then a temporary directory will be used that is deleted after data import.
catch_output (bool, optional) – if True, do not show CRTomo output. Default:True
cores (int, optional) – how many cores to use for CRTomo
**kwargs (Are supplied to
crtomo.tdMan._invert())
- Returns:
return_code – Return 0 if the inversion completed successfully. Return 1 if no measurements are present.
- Return type:
- load_rho_file(filename)[source]¶
Load a forward model from a rho.dat file
- Parameters:
filename (string) – filename to rho.dat file
- Returns:
pid_mag (int) – parameter id for the magnitude model
pid_pha (int) – parameter id for the phase model
- measurements(silent=False)[source]¶
Return the measurements associated with this instance.
If measurements are not present, check if we can model, and then run CRMod to load the measurements.
- Parameters:
silent (bool, optional) – If False, suppress CRMod output
- model(voltages=True, sensitivities=False, potentials=False, output_directory=None, silent=False)[source]¶
Forward model the tomodir and read in the results
Note that this function will always do the full modeling. No caching involved.
Please use .measurements to accessed cached measurements.
- Parameters:
voltages (bool, optional) – if True, compute voltages for registered quadrupoles. Default: True
sensitivities (bool, optional) – if True, compute sensitivities for registered quadrupoles. Default: False
potentials (bool, optional) – if True, compute potential fields for all current injections. Default: False TODO: check if implemented in the Python wrapper
output_directory (str|None, optional) – if this is a string, treat it as an output directory in which the tomodir used for the modeling is saved. Will raise an exception if the output directory already exists. Default: None
silent (bool, optional) – if True, suppress most of the output. Default: False
- Return type:
None
- plot_decouplings_to_ax(ax, plot_transistions=True)[source]¶
Visualize decouplings. Usually you may want to plot the mesh to this axis first
- Parameters:
ax (matplotlib.Axes) – Axis to plot to
plot_transistions (bool (True)) – If True, then also visualize the decoupled transitions
- plot_eps_data(filename=None)[source]¶
Plot data residuals and data error weighting
- Parameters:
filename (string|None) – if not None, then save plot to this file
- Returns:
figure – if filename is None, then return the generated figure
- Return type:
matplotlib.Figure|None
- plot_eps_data_hist(filename=None)[source]¶
Plot histograms of data residuals and data error weighting
- TODO :
add percentage of data below/above the RMS value
- Parameters:
filename (string|None) – if not None, then save plot to this file
- Returns:
figure – if filename is None, then return the generated figure
- Return type:
matplotlib.Figure|None
- plot_forward_models(mag_only=False, separate_figures=False, **kwargs)[source]¶
Plot the forward models (requires the forward models to be loaded)
- plot_inversion_evolution(filename=None)[source]¶
Plot the evolution of inversion properties (lambda, RMS, …) :param filename: if not None, save plot to this file :type filename: string|None
- Returns:
fig – if filename is None, return the figure object
- Return type:
matplotlib.Figure|None
- plot_inversion_result_rmag(figsize=None, log10=False, overlay_coverage=False, **kwargs)[source]¶
Plot the final inversion results, magnitude-results only
- Parameters:
figsize ((float, float)|None) – Figure size of the matplotlib figure in inches.
log10 (bool, default: False) – Shortcut to force a log10 conversion of the resistivity data
overlay_coverage (bool, default: False) – If True, use the cumulated coverage to adjust the transparency of the plot
**kwargs (dict) – will be propagated into self.plot.plot_elements_to_ax
- Returns:
fig (matplotlib.Figure) – The created figure
ax (matplotlib.Axes) – Plot axes
- plot_pseudo_locs(spacing=1.0)[source]¶
Plot pseudo-locations of measurement configurations. This function does not take into account real electrode locations. However, for surface configurations a ‘spacing’ parameter can be provided
- Parameters:
spacing (float, default=1.0) – Electrode spacing
- plot_sensitivity(config_nr=None, sens_data=None, mag_only=False, absv=False, **kwargs)[source]¶
Create a nice looking plot of the sensitivity distribution for the given configuration nr. Configs start at 1!
- Parameters:
config_nr (int, optional) – The configuration number (starting with 0) to compute the sensitivity for.
sens_data (Nx2 numpy.ndarray, optional) – If provided, use this data as sensitivity data (do not compute anything)
mag_only (bool, optional) – Plot only the magnitude sensitivities
absv (bool, optional) – If true, plot absolute values of sensitivity
- Returns:
fig
ax
Examples
(
Source code,png,hires.png,pdf)
- read_decouplings_file(filename)[source]¶
Import decoupling data for the inversion. This is usally a file called decouplings.dat in the exe/ directory of a tomodir, but we can also read from an BytesIO object.
Do nothing if the file does not exist
Overwrite any existing decouplings
- read_inversion_results(tomodir)[source]¶
Import inversion results from a tomodir into this instance
Warning
This function is not finished yet and does not import ALL crtomo information yet.
- Parameters:
tomodir (str) – Path to tomodir
- read_voltages(voltage_file)[source]¶
Import voltages from a volt.dat file
- Parameters:
voltage_file (str) – Path to volt.dat file
- register_data_errors(mid_rmag_error, mid_rpha_error=None, norm_mag=1, norm_pha=1)[source]¶
Register individual data errors.
The normalization factors are used as follows:
mag_error -> mag_error / norm_mag ** 2 pha_error -> pha_error / norm_pha ** 2
That means that you need to decrease the values in order to increase individual errors.
- Parameters:
mid_mag_error (int) – ID to the corresponding measurement in tdMan.configs holding the magnitude errors (linear)
mid_pha_error (int) – ID to the corresponding measurement in tdMan.configs holding the phase errors (linear)
norm_mag (float) – Magnitude errors can be normalized by the square of this value.
norm_pha (float) – Phase errors can be normalized by the square of this value.
- register_forward_model(pid_mag, pid_pha)[source]¶
Register parameter sets as the forward models for magnitude and phase
- register_measurements(mag, pha=None)[source]¶
Register measurements as magnitude/phase measurements used for the inversion
- Parameters:
mag (int|numpy.ndarray) – magnitude measurement id for the corresponding measurement data in self.configs.measurements. If mag is a numpy.ndarray, assume mag to be the data itself an register it
pha (int, optional) – phase measurement id for the corresponding measurement data in self.configs.measurements. If not present, a new measurement set will be added with zeros only.
- reset_data()[source]¶
Attempts to reset (delete) all inversion data currently stored in the tdMan instance. This is mostly attempted for the impedance data (magnitudes, phases, conductivity real and imaginary parts), but could be extended to other data (this is currently not done due to complexity and missing demand). Forward models are also deleted.
- save_measurements(filename)[source]¶
Save measurements in a file. Use the CRTomo format.
Do not save anything if no measurements are present in this tdMan instance.
- Parameters:
filename (string) – path to output filename
- save_to_tarfile()[source]¶
Save the current modeling/inversion data into a tarfile. Return the file as an io.BytesIO object. The tar file is generated completely in memory - no files are written to the disc
- Returns:
tomodir_tar – Tomodir stored in tar file
- Return type:
- save_to_tomodir(directory, only_for_inversion=False)[source]¶
Save the tomodir instance to a directory structure.
At this point forward modeling results (voltages, potentials and sensitivities) will be saved.
Inversion results will, at this stage, not be saved (TODO!!!).
- Parameters:
directory (str) – Output directory (tomodir). Will be created of it does not exists, but otherwise files we be overwritten in any existing directories
only_for_inversion (bool, optional) – If True, save only files required for an inversion (i.e., omit any forward modeling files and results not necessary). Default: False
Note
Test cases:
modeling only
inversion only
modeling and inversion
- show_parset(pid, **kwargs)[source]¶
Plot a given parameter set. Thin wrapper around
crtomo.plotManager.plotManager.plot_elements_to_ax().kwargs will be directly supplied to the plot function
Module contents¶
- class crtomo.ConfigManager(**kwargs)[source]¶
Bases:
ConfigManager- _crmod_to_abmn(configs)[source]¶
convert crmod-style configurations to a Nx4 array
CRMod-style configurations merge A and B, and M and N, electrode numbers into one large integer each:
\[ \begin{align}\begin{aligned}AB = A \cdot 10^4 + B\\MN = M \cdot 10^4 + N\end{aligned}\end{align} \]- Parameters:
configs (numpy.ndarray) – Nx2 array holding the configurations to convert
Examples
>>> import numpy as np >>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=5) >>> crmod_configs = np.array(( ... (10002, 40003), ... (10010, 30004), ... )) >>> abmn = config._crmod_to_abmn(crmod_configs) >>> print(abmn) [[ 1 2 4 3] [ 1 10 3 4]]
- _get_crmod_abmn()[source]¶
return a Nx2 array with the measurement configurations formatted CRTomo style
- abmn_to_dataframe()[source]¶
Return a pandas.DataFrame containing the measurement configurations
- Returns:
abmn_df – Configurations in a DataFrame (columns: a,b,m,n)
- Return type:
- add_measurements(measurements)[source]¶
Add new measurements to this instance
- Parameters:
measurements (numpy.ndarray) – one or more measurement sets. It must either be 1D or 2D, with the first dimension the number of measurement sets (K), and the second the number of measurements (N): K x N
- Returns:
mid – measurement ID used to extract the measurements later on
- Return type:
Examples
>>> import numpy as np import crtomo.configManager as CRconfig config = CRconfig.ConfigManager(nr_of_electrodes=10) config.gen_dipole_dipole(skipc=0) # generate some random noise random_measurements = np.random.random(config.nr_of_configs) mid = config.add_measurements(random_measurements) # retrieve using mid print(config.measurements[mid])
- add_to_configs(configs)[source]¶
Add one or more measurement configurations to the stored configurations
- Parameters:
configs (list or numpy.ndarray) – list or array of configurations
- Returns:
configs – array holding all configurations of this instance
- Return type:
Kx4 numpy.ndarray
- clear_measurements()[source]¶
Remove all measurements from self.measurements. Reset the measurement counter. All ID are invalidated.
- delete_data_points(indices)[source]¶
Delete data points by index (0-indexed), both in configs and measurements. Deletions will be done in ALL registered measurements to ensure consistency.
- Parameters:
indices (int|iterable) – Indices to delete
- gen_all_current_dipoles()[source]¶
Generate all possible current dipoles for the given number of electrodes (self.nr_electrodes). Duplicates are removed in the process.
After Noel and Xu, 1991, for N electrodes, the number of possible unique configurations is \(N \cdot (N - 1) / 2\). This excludes duplicates in the form of switches current/voltages electrodes, as well as reciprocal measurements.
- Returns:
configs – all possible current dipoles A-B
- Return type:
Nx2 numpy.ndarray
- gen_all_voltages_for_injections(injections_raw)[source]¶
For a given set of current injections AB, generate all possible unique potential measurements.
After Noel and Xu, 1991, for N electrodes, the number of possible voltage dipoles for a given current dipole is \((N - 2)(N - 3) / 2\). This includes normal and reciprocal measurements.
If current dipoles are generated with ConfigManager.gen_all_current_dipoles(), then \(N \cdot (N - 1) / 2\) current dipoles are generated. Thus, this function will produce \((N - 1)(N - 2)(N - 3) / 4\) four-point configurations ABMN, half of which are reciprocals (Noel and Xu, 1991).
All generated measurements are added to the instance.
Use ConfigManager.split_into_normal_and_reciprocal() to split the configurations into normal and reciprocal measurements.
- Parameters:
injections (numpy.ndarray) – Kx2 array holding K current injection dipoles A-B
- Returns:
configs – Nax4 array holding all possible measurement configurations
- Return type:
- gen_configs_permutate(injections_raw, only_same_dipole_length=False, ignore_crossed_dipoles=False, silent=False)[source]¶
Create measurement configurations out of a pool of current injections. Use only the provided dipoles for potential dipole selection. This means that we have always reciprocal measurements.
Remove quadpoles where electrodes are used both as current and voltage dipoles.
- Parameters:
injections_raw (Nx2 array) – current injections
only_same_dipole_length (bool, optional) – if True, only generate permutations for the same dipole length
ignore_crossed_dipoles (bool, optional) – If True, potential dipoles will be ignored that lie between current dipoles, e.g. 1-4 3-5. In this case it is possible to not have full normal-reciprocal coverage.
silent (bool, optional) – if True, do not print information on ignored configs (default: False)
- Returns:
configs – quadrupoles generated out of the current injections
- Return type:
Nx4 array
- gen_dipole_dipole(skipc, skipv=None, stepc=1, stepv=1, nr_voltage_dipoles=10, before_current=False, allow_crossings=False, start_skip=0, N=None)[source]¶
Generate dipole-dipole configurations
- Parameters:
skipc (int) – number of electrode positions that are skipped between electrodes of a given dipole
skipv (int) – steplength between subsequent voltage dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
stepc (int) – steplength between subsequent current dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
stepv (int) – steplength between subsequent voltage dipoles. A steplength of 0 will produce increments by one, i.e., 3-4, 4-5, 5-6 …
nr_voltage_dipoles (int) – the number of voltage dipoles to generate for each current injection dipole
before_current (bool, optional) – if set to True, also generate voltage dipoles in front of current dipoles.
allow_crossings (bool, optional) – Allow potential dipoles to cross current injection dipoles. Note that any quadpole generated by crossing the current electrodes is NOT counted into nr_voltage_dipoles.
start_skip (int, optional) – how many electrode to skip before/after the first/second current electrode.
N (int, optional) – number of electrodes, must be given if not already known by the config instance
Examples
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole(skipc=2) array([[ 1, 4, 5, 8], [ 1, 4, 6, 9], [ 1, 4, 7, 10], [ 2, 5, 6, 9], [ 2, 5, 7, 10], [ 3, 6, 7, 10]])
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole( ... skipc=2, before_current=True, allow_crossings=True) array([[ 1, 4, 2, 5], [ 1, 4, 3, 6], [ 1, 4, 5, 8], [ 1, 4, 6, 9], [ 1, 4, 7, 10], [ 2, 5, 1, 4], [ 2, 5, 3, 6], [ 2, 5, 4, 7], [ 2, 5, 6, 9], [ 2, 5, 7, 10], [ 3, 6, 1, 4], [ 3, 6, 2, 5], [ 3, 6, 4, 7], [ 3, 6, 5, 8], [ 3, 6, 7, 10]])
>>> from reda.configs.configManager import ConfigManager >>> config = ConfigManager(nr_of_electrodes=10) >>> config.gen_dipole_dipole(skipc=1, before_current=True) array([[ 1, 3, 4, 6], [ 1, 3, 5, 7], [ 1, 3, 6, 8], [ 1, 3, 7, 9], [ 1, 3, 8, 10], [ 2, 4, 5, 7], [ 2, 4, 6, 8], [ 2, 4, 7, 9], [ 2, 4, 8, 10], [ 3, 5, 6, 8], [ 3, 5, 7, 9], [ 3, 5, 8, 10], [ 4, 6, 1, 3], [ 4, 6, 7, 9], [ 4, 6, 8, 10], [ 5, 7, 2, 4], [ 5, 7, 1, 3], [ 5, 7, 8, 10]])
- gen_inner_gradients(injections, skips=None)[source]¶
Given a set of current injections, generate voltage dipoles between the current electrodes
Examples
>>> import reda >>> cfg = reda.ConfigManager(nr_of_electrodes=10) >>> cfg.gen_inner_gradients([1, 10], skips=[2, ]) array([[ 1, 10, 2, 5], [ 1, 10, 3, 6], [ 1, 10, 4, 7], [ 1, 10, 5, 8], [ 1, 10, 6, 9]])
- Parameters:
injections (Nx2 numpy.ndarray) – Current injections
skips (None or iterable) – The skips to generate. None will generate all possible skips
- Returns:
configs – The generated configurations
- Return type:
Nx4 numpy.ndarray
- gen_reciprocals(append=False)[source]¶
Generate reciprocal configurations, sort by AB, and optionally append to configurations.
- Parameters:
append (bool) – Append reciprocals to configs (the default is False).
Examples
>>> cfgs = ConfigManager(nr_of_electrodes=5) >>> nor = cfgs.gen_dipole_dipole(skipc=0) >>> rec = cfgs.gen_reciprocals(append=True) >>> print(cfgs.configs) [[1 2 3 4] [1 2 4 5] [2 3 4 5] [3 4 1 2] [4 5 1 2] [4 5 2 3]]
- gen_schlumberger(M, N, a=None)[source]¶
generate one Schlumberger sounding configuration, that is, one set of configurations for one potential dipole MN.
- Parameters:
- Returns:
configs – array holding the configurations
- Return type:
Kx4 numpy.ndarray
Examples
import reda.configs.configManager as CRconfig config = CRconfig.ConfigManager(nr_of_electrodes=40) config.gen_schlumberger(M=20, N=21)
- gen_voltage_dipoles_skip(cinj, skip=0)[source]¶
For given current injections, generate all possible voltage dipoles with skip skip.
- Parameters:
cinj (
numpy.ndarray) – Nx2 array containing the current injection electrodes a,bskip (int, optional) – Skip used for voltage electrodes. Default: 0
- gen_wenner(a)[source]¶
Generate Wenner measurement configurations.
- Parameters:
a (int) – distance (in electrodes) between subsequent electrodes of each four-point configuration.
- Returns:
configs – array holding the configurations
- Return type:
Kx4 numpy.ndarray
- get_unique_current_injections()[source]¶
Return all unique current injection electrode pairs
Examples
>>> from reda.configs.configManager import ConfigManager >>> cfg = ConfigManager() >>> cfg.nr_electrodes >>> cfg.nr_electrodes = 10 >>> cfg.add_to_configs(( ... (1, 2, 3, 4), ... (1, 2, 5, 6), ... (7, 8, 9, 10) ... )) array([[ 1, 2, 3, 4], [ 1, 2, 5, 6], [ 7, 8, 9, 10]]) >>> cfg.get_unique_current_injections() array([[1, 2], [7, 8]])
cfg = ConfigManager() cfg.get_unique_current_injections()
- Returns:
unique_injections – Current injections
- Return type:
Nx2 numpy.ndarray
- load_crmod_config(filename)[source]¶
Load a CRMod configuration file
- Parameters:
filename (string|io.BytesIO) – absolute or relative path to a crmod config.dat file. Can also be a BytesIO object
- load_crmod_data(data_source, is_forward_response=False, try_fix_signs=False)[source]¶
Load CRMod measurement data, either from file or directly from a numpy array.
- Parameters:
data_source (string|numpy.ndarray) – if this is a string, treat it as an input filename. If it is a Nx6 or Nx3 numpy array, use this data directly
is_forward_response (bool, optional) – If True this indicates a volt.dat file created by CRTomo during an inversion run to store forward responses of a given model iteration. In this case the third data column indicates the wdfak parameter, i.e., it indicates if a given data point was excluded from this iteration.
try_fix_signs (bool, optional) – If True, try to fix sign reversals with respect to an already registered configuration set by swappend m and n entries.
- Returns:
cid_mag (int) – Measurement id for magnitude data
cid_pha (int) – Measurement id for phase data
mid_wdfak (int, optional) – Measurement id for wdfak indicators. Only returned if is_forward_response is True
- load_crmod_or_4c_configs(filename)[source]¶
Load configurations either from a CRMod measurement file, or from a four-column file. Assume 1-indexed data (start with electrode 1).
- Parameters:
filename (str) – Path to file that shall be imported
- Return type:
None
- load_crmod_volt(filename)[source]¶
Load a CRMod measurement file (commonly called volt.dat)
- Parameters:
filename (string) – path to input filename
- Returns:
cid_mag (int) – Measurement id for magnitude data
cid_pha (int) – Measurement id for phase data
- load_injections_from_mcf(filename)[source]¶
Load injections from a mcf-file used by the sEIT-systems from FZJ; Injections in the file must be formatted like the following: SE 001 002 SE 002 001 . . .
- Parameters:
filename (string) – absolute or relative path to a mcf-file
- Return type:
Nx2 numpy array with current injections
- property nr_of_configs¶
Return number of configurations
- Returns:
nr_of_configs – number of configurations stored in this instance
- Return type:
- remove_duplicates(configs=None)[source]¶
remove duplicate entries from 4-point configurations. If no configurations are provided, then use self.configs. Unique configurations are only returned if configs is not None.
- Parameters:
configs (Nx4 numpy.ndarray, optional) – remove duplicates from these configurations instead from self.configs.
- Returns:
configs_unique – unique configurations. Only returned if configs is not None
- Return type:
Kx4 numpy.ndarray
- remove_max_dipole_sep(maxsep=10)[source]¶
Remove configurations with dipole separations higher than maxsep.
- Parameters:
maxsep (int) – Maximum separation between both dipoles (the default is 10).
- remove_using_nr_injections(min_nr_of_injections)[source]¶
Remove all measurements with a current injection that is not used a minimum number of times.
This is useful to optimize measurement time for multi-channel systems, such as the IRIS Instruments Syscal Pro. The device can theoretically only facilitate the full number of channels if a corresponding number of voltage measurements is requested for a given current injection. As such, it can be useful to remove measurements with unique current injection dipoles. Note that other factors determine if all channels are actually used. Please refer to the device manual for device-specific information.
- Parameters:
min_nr_of_injections (int) – Minimum number a given current injection should have to keep it
- split_into_normal_and_reciprocal(pad=False, return_indices=False)[source]¶
Split the stored configurations into normal and reciprocal measurements
** Rule 1: the normal configuration contains the smallest electrode number of the four involved electrodes in the current dipole **
- Parameters:
- Returns:
normal (numpy.ndarray) – Nnx4 array. If pad is True, then Nn == N (total number of unique measurements). Otherwise Nn is the number of normal measurements.
reciprocal (numpy.ndarray) – Nrx4 array. If pad is True, then Nr == N (total number of unique measurements). Otherwise Nr is the number of reciprocal measurements.
nor_indices (numpy.ndarray, optional) – Nnx1 array containing the indices of normal measurements. Only returned if return_indices is True.
rec_indices (numpy.ndarray, optional) – Nrx1 array containing the indices of normal measurements. Only returned if return_indices is True.
- to_iris_syscal(filename, spacing=1)[source]¶
Export to IRIS Instrument configuration file
- Parameters:
filename (string) – Path to output filename
- to_pg_scheme(container=None, positions=None)[source]¶
Convert the configuration to a pygimli measurement scheme
- Parameters:
container (reda.containers.ERT.ERT) – an ERT data container (we take the electrode positions from here)
None (positions =)
- Returns:
data
- Return type:
pybert.DataContainerERT
Examples
import numpy as np from reda.configs.configManager import ConfigManager configs = ConfigManager(nr_of_electrodes=48) new_configs = configs.gen_dipole_dipole(skipc=2) x = np.arange(0, 48, 1) z = np.ones(48) * -1 y = np.zeros(48) xyz = np.vstack((x, y, z)).T scheme = configs.to_pg_scheme(positions=xyz) print(scheme)
- property unique_injections¶
- write_crmod_config(filename)[source]¶
Write the configurations to a configuration file in the CRMod format All configurations are merged into one previor to writing to file
- Parameters:
filename (string) – absolute or relative path to output filename (usually config.dat)
- write_crmod_volt(filename, mid)[source]¶
Write the measurements to the output file in the volt.dat file format that can be read by CRTomo.
Data is written in utf-8 encoding.
- Parameters:
filename (string|io.BytesIO) – output filename
mid (int or [int, int]) – measurement ids of magnitude and phase measurements. If only one ID is given, then the phase column is filled with zeros
- write_crmod_volt_with_individual_errors(filename, data_mids, error_mids, norm_mag=1, norm_pha=1)[source]¶
Write the measurements and individual errors to the output file in the volt.dat file format that can be read by CRTomo.
Data is written in utf-8 encoding.
- Parameters:
filename (string|io.BytesIO) – output filename
data_mids ([int, int]) – measurement ids of magnitude and phase measurements
error_mids ([int, int]) – ids of magnitude and phase error estimates
norm_mag (float) – Normalization factor for magnitude errors
norm_pha (float) – Normalization factor for magnitude errors
- class crtomo.ParMan(grid_obj)[source]¶
Bases:
objectmanage one or more parameter sets for a given
crtomo.grid.crt_gridobject- __init__(grid_obj)[source]¶
- Parameters:
grid_obj (crtomo.grid.crt_grid) – The FE grid that the parameter sets refer to
- _clean_pid(pid)[source]¶
if pid is a number, don’t do anything. If pid is a list with one entry, strip the list and return the number. If pid contains more than one entries, do nothing.
- add_2d_cos_anomaly_line(pid, p0, anomaly_width, anomaly_height, peak_value, area='only_one_y', only_one_y_line=True, only_one_x_line=False, whole_mesh=False)[source]¶
Add one line of cos(x)cos(y) anomalies to a given parameter set. The wavelength refers to half a period, with the maximum of the anomaly centered on p0=[x0, z0].
- Parameters:
whole_mesh (bool, optional) – If True, then fill the whole mesh with a cos(x)cos(y) pattern.
- add_checkerboard_pattern(pid, p0, anomaly_width, anomaly_height, peak_value)[source]¶
Note that if p0 is larger than a few anomaly sizes its possible that the checkerboard pattern will only be applied to parts of the mesh!
- add_data(data, metadata=None)[source]¶
Add data to the parameter set
- Parameters:
data (numpy.ndarray) – one or more parameter sets. It must either be 1D or 2D, with the first dimension the number of parameter sets (K), and the second the number of elements (N): K x N
metadata (object, optional) – the provided object will be stored in in the metadata dict and can be received with the ID that is returned. If multiple (K) datasets are added at ones, provide a list of objects with len K.
- Returns:
ID which can be used to access the parameter set
- Return type:
int, ID
Examples
>>> # suppose that grid is a fully initialized grid oject with 100 # elements parman = ParMan(grid) # one_data_set = np.ones(100) cid = parman.add_data(one_data_set) print(parman.parsets[cid]) two_data_sets = np.ones((2, 100)) cids = parman.add_data(two_data_sets) print(cids) [0, ] [1, 2]
- add_empty_dataset(value=1)[source]¶
Create an empty data set. Empty means: all elements have the same value.
- Parameters:
value (float, optional) – which value to assign all element parameters. Default is one.
- add_gaussian_anomaly_to_parset(pid, center, width, max_value)[source]¶
- center_of_mass_value(pid, mode='log10')[source]¶
Compute the center of mass value of a given parameter set.
- create_parset_with_gaussian_anomaly(center, width, max_value, background)[source]¶
- Parameters:
- Returns:
pid – ID of newly created parameter set
- Return type:
- extract_along_line(pid, xy0, xy1, N=10)[source]¶
Extract parameter values along a given line.
- Parameters:
- Returns:
values – data values for extracted data points. First column: x-coordinates, second column: z-coordinates, third column: extracted values
- Return type:
numpy.ndarray (n x 3)
- extract_points(pid, points)[source]¶
Extract values at certain points in the grid from a given parameter set. Cells are selected by interpolating the centroids of the cells towards the line using a “nearest” scheme.
Note that data is only returned for the points provided. If you want to extract multiple data points along a line, defined by start and end point, use the extract_along_line function.
- Parameters:
pid (int) – The parameter id to extract values from
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
values – data values for extracted data points
- Return type:
numpy.ndarray (n x 1)
- extract_polygon_area(pid, polygon_points)[source]¶
Extract all data points whose element centroid lies within the given polygon.
- Parameters:
- Returns:
elements_in_polygon_array (numpy.ndarray) – list with elements numbers (zero-indexed, starting from 0) which lie within the polygon
values (list) – the data values corresponding to the elements in elements_in_polygon
- load_from_rho_file(filename)[source]¶
Convenience function that loads two parameter sets from a rho.dat file, as used by CRMod for forward resistivity/phase models.
- Parameters:
filename (string, file path) – filename of rho.dat file
- Returns:
cid_mag (int) – ID of magnitude parameter set
cid_phase (int) – ID of phase parameter set
- load_from_sens_file(filename)[source]¶
Load real and imaginary parts from a sens.dat file generated by CRMod
- Parameters:
filename (string) – filename of sensitivity file
- Returns:
nid_re (int) – ID of real part of sensitivities
nid_im (int) – ID of imaginary part of sensitivities
- load_inv_result(filename, columns=2, is_log10=False)[source]¶
Load one parameter set from a rho*.mag or rho*.pha file produced by CRTomo.
- Parameters:
- Returns:
pid – ID(s) of parameter set
- Return type:
- load_model_from_file(filename, columns=0)[source]¶
Load one parameter set from a file which contains one value per line
No row is skipped.
- modify_area(pid, xmin, xmax, zmin, zmax, value)[source]¶
Modify the given dataset in the rectangular area given by the parameters and assign all parameters inside this area the given value.
Partially contained elements are treated as INSIDE the area, i.e., they are assigned new values.
- Parameters:
pid (int) – id of the parameter set to modify
xmin (float) – smallest x value of the area to modify
xmax (float) – largest x value of the area to modify
zmin (float) – smallest z value of the area to modify
zmin – largest z value of the area to modify
value (float) – this value is assigned to all parameters of the area
Examples
>>> import crtomo.tdManager as CRtdm tdman = CRtdm.tdMan( elem_file='GRID/elem.dat', elec_file='GRID/elec.dat', ) pid = tdman.parman.add_empty_dataset(value=1) tdman.parman.modify_area( pid, xmin=0, xmax=2, zmin=-2, zmin=-0.5, value=2, ) fig, ax = tdman.plot.plot_elements_to_ax(pid) fig.savefig('out.png')
- modify_polygon(pid, polygon, value)[source]¶
Modify parts of a parameter set by modifying all elements within a provided
shapely.geometry.Polygoninstance. Hereby, an element is modified if its center lies within the polygon.- Parameters:
Examples
>>> import shapely.geometry polygon = shapely.geometry.Polygon(( (2, 0), (4, -1), (2, -1) )) tdman.parman.modify_polygon_centroids(pid, polygon, 3)
- modify_polygon_old(pid, polygon, value)[source]¶
Modify parts of a parameter set by setting all parameters located in, or touching, the provided
shapely.geometry.Polygoninstance.WARNING: This implementation often leads to ragged borders in the selected polygons. Use the new modify_polygon function!
- Parameters:
Examples
>>> import shapely.geometry polygon = shapely.geometry.Polygon(( (2, 0), (4, -1), (2, -1) )) tdman.parman.modify_polygon(pid, polygon, 3)
- reset()[source]¶
Resets the ParMan instance. This process deletes all data and metadata, and resets the index variable
- class crtomo.crt_grid(elem_file=None, elec_file=None, empty=False)[source]¶
Bases:
objectThe
crt_gridholds and manages one Finite-Element grid.This class provides only basic plotting routines. Advanced plotting routines can be found in
crtomo.plotManager.Examples
>>> import crtomo.grid as CRGrid grid = CRGrid(elem_file='elem.dat', elec_file='elec.dat') print(grid) CRMod/CTRomo grid instance number of elements: 2728 number of nodes: 1440 number of electrodes: 60 grid dimsensions: X: -7.5 37.5 Z: -15.0 0.0
- Wm()[source]¶
Return the smoothing regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.48ff
\[\begin{split}\Psi_m = ||\underline{\underline{W}}_m \underline{m}||^2_2\\ = \sum_{j=1}^M \sum_{i=nb(j)} \alpha_{r_ij} |\frac{m_j - m_i}{\Delta c_{ij}|^2 \Delta b_{ij} \Delta c_{ij}\end{split}\]Note that the anisotropic regularization parameter \(\alpha\) is not implemented yet.
i and j are swapped in the implementation.
See Fig 3.5 in the thesis.
- Wm_mgs(m, beta)[source]¶
Return the MGS regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.50
Note that alpha and f are set 1
- Parameters:
m (numpy.ndarray) – model parameters to compute MGS matrix for
beta (float) – MGS beta parameter
- Returns:
Wm – Sparse MGS matrix
- Return type:
- static _get_area_polygon(points_x, points_z)[source]¶
Return the area of a polygon. The node coordinates must be ordered, but the direction does not matter - we return the abs-value
>>> points_x = [4, 4, 8, 8, -4, -4] >>> points_z = [6, -4, -4, -8, -8, 6] >>> self._get_area_polygon(points_x, points_z) ... 128
- _prepare_grids()[source]¶
depending on the type of grid (rectangular or triangle), prepare grids or triangle lists
- TODO: We want some nice way of not needing to know in the future if we
loaded triangles or quadratic elements.
- _read_elem_elements(fid)[source]¶
Read all FE elements from the file stream. Elements are stored in the self.element_data dict. The keys refer to the element types:
3: Triangular grid (three nodes)
8: Quadrangular grid (four nodes)
11: Mixed boundary element
12: Neumann (no-flow) boundary element
- _read_elem_nodes(fid)[source]¶
Read the nodes from an opened elem.dat file. Correct for CutMcK transformations.
We store three typed of nodes in the dict ‘nodes’:
“raw” : as read from the elem.dat file
“presort” : pre-sorted so we can directly read node numbers from a elec.dat file and use them as indices.
- “sorted”completely sorted as in the original grid (before any
CutMcK)
For completeness, we also store the following keys:
“cutmck_index” : Array containing the indices in “presort” to obtain the “sorted” values: nodes[‘sorted’] = nodes[‘presort’] [nodes[‘cutmck_index’], :]
“rev_cutmck_index” : argsort(cutmck_index)
- analyze_internal_angles(return_plot=False)[source]¶
Analyze the internal angles of the grid. Angles shouldn’t be too small because this can cause problems/uncertainties in the Finite-Element solution of the forward problem. This function prints the min/max values, as well as quantiles, to the command line, and can also produce a histogram plot of the angles.
- Parameters:
return_plot (bool) – if true, return (fig, ax) objects of the histogram plot
- Returns:
fig (matplotlib.figure) – figure object
ax (matplotlib.axes) – axes object
Examples
>>> import crtomo.grid as CRGrid grid = CRGrid.crt_grid() grid.load_elem_file('elem.dat') fig, ax = grid.analyze_internal_angles(Angles) This grid was sorted using CutMcK. The nodes were resorted! Triangular grid found Minimal angle: 22.156368696965796 degrees Maximal angle: 134.99337326279496 degrees Angle percentile 10%: 51.22 degrees Angle percentile 20%: 55.59 degrees Angle percentile 30%: 58.26 degrees Angle percentile 40%: 59.49 degrees Angle percentile 50%: 59.95 degrees Angle percentile 60%: 60.25 degrees Angle percentile 70%: 61.16 degrees Angle percentile 80%: 63.44 degrees Angle percentile 90%: 68.72 degrees generating plot... >>> # save to file with fig.savefig('element_angles.png', dpi=300)
- static create_surface_grid(nr_electrodes=None, spacing=None, electrodes_x=None, electrodes_z=None, depth=None, left=None, right=None, char_lengths=None, lines=None, internal_lines=None, debug=False, workdir=None, force_neumann_only=False)[source]¶
This is a simple wrapper for cr_trig_create to create simple surface grids.
Electrode and boundary positions are rounded to the third digit.
- Parameters:
nr_electrodes (int, optional) – the number of surface electrodes
spacing (float, optional) – the spacing between electrodes, usually in [m], required if nr of electrodes is given
electrodes_x (array, optional) – x-electrode positions can be provided here, e.g., for non-equidistant electrode distances
electrodes_z (array, optional) – z-electrode positions can be provided here, e.g., for non-equidistant electrode distances. Only useful in combination with electrodes_x
depth (float, optional) – the depth of the grid, relative to the minimum z-value. If not given, this is computed as half the maximum distance between electrodes
left (float, optional) – the space allocated left of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
right (float, optional) – the space allocated right of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
char_lengths (float|list of 4 floats, optional) – characteristic lengths, as used by cr_trig_create
lines (list of floats, optional) – at the given depths, add horizontal lines in the grid. Note that all positive values will be multiplied by -1!
internal_lines (list of 4-tuple) – extra lines to add to the grid. Important: These lines must NOT touch the outer edge of the grid (this is not supported by this function, but can be accomplished by manually building the grid)
debug (bool, optional) – default: False. If true, don’t hide the output of cr_trig_create
workdir (string, optional) – if set, use this directory to create the grid. Don’t delete files afterwards.
force_neumann_only (bool, optional) – sometimes we want to use only Neumann boundary conditions. Setting this switch to True will force all boundaries to Neumann. Use only if you know what you are doing! default: False
- Returns:
grid – the generated grid
- Return type:
crtomo.grid.crt_gridinstance
Examples
>>> from crtomo.grid import crt_grid >>> grid = crt_grid.create_surface_grid(40, spacing=0.25, depth=5, ... left=2, right=2, char_lengths=[0.1, 0.5, 0.1, 0.5], ... lines=[0.4, 0.8], debug=False, workdir=None) >>> import pylab as plt >>> fig, ax = plt.subplots() >>> grid.plot_grid_to_ax(ax)
- property element_neighbors¶
- property element_neighbors_old¶
Return a list with element numbers (zero indexed) of neighboring elements. Note that the elements are not sorted. No spacial orientation can be inferred from the order of neighbors.
WARNING: This function is slow due to a nested loop. This would be a good starting point for further optimizations.
In order to speed things up, we could search using the raw data, i.e., with CutMcK enabled sorting, and then restrict the loops to 2x the bandwidth (before - after).
While not being returned, this function also sets the variable self.element_neighbors_edges, in which the common nodes with each neighbor are stored.
- Returns:
neighbors – a list (length equal to nr of elements) with neighboring elements
- Return type:
Examples
- get_electrode_node(electrode)[source]¶
For a given electrode (e.g. from a config.dat file), return the true node number as in self.nodes[‘sorted’]
- get_electrode_positions()[source]¶
Return the electrode positions in an numpy.ndarray
- Returns:
positions – Nx2 array, where N is the number of electrodes. The first column contains x positions, the second z positions.
- Return type:
- get_element_areas()[source]¶
return the areas of the elements
note that this formula is generic, see CRMod code for a simpler one
- Returns:
areas
- Return type:
- get_element_centroids()[source]¶
return the central points of all elements
- Returns:
centroids – Nx2 array x/z coordinates for all (N) elements
- Return type:
- get_element_indices_within_rectangle(xmin, xmax, zmin, zmax)[source]¶
Return the indices of all elements whose center is located within the rectangle defined by the parameters.
The indices can then be used, e.g., to select values from inversion results.
- Parameters:
- Returns:
indices – Array with indices (zero-indexed)
- Return type:
np.array
- get_internal_angles()[source]¶
Compute all internal angles of the grid
- Returns:
NxK array with N the number of elements, and K the number of nodes, filled with the internal angles in degrees
- Return type:
- get_min_max_electrode_distances()[source]¶
Return the minimal and the maximal electrode distance of the grid
- Returns:
amin (float) – minimal electrode distance
amax (float) – maximal electrode distance
- get_minmax()[source]¶
Return min/max x/z coordinates of grid
- Returns:
x ([float, float]) – min, max values of grid dimensions in x direction (sideways)
z ([float, float]) – min, max values of grid dimensions in z direction (downwards)
- static interpolate_grid(ingrid, outgrid, data, method='nearest')[source]¶
Function for interpolating data from one grid to another, using the cell midpoints as datapoint locations. Standard method for interpolating is set to nearest-neighbour.
- Parameters:
ingrid (
crtomo.grid.crt_gridinstance) – CRT grid that matches the input dataoutgrid (
crtomo.grid.crt_gridinstance) – CRT grid to interpolate todata (pandas.dataframe) – input data that matches the input grid (in pandas dataframe format)
method (interpolation method, optional) – Standard interpolation method is nearest-neighbour (‘nearest’). Other possible methods are ‘linear’ and ‘cubic’.
- Returns:
interpolated data – returned dataframe with interpolated data
- Return type:
pandas.dataframe
- load_elem_file(elem_file)[source]¶
Load a CRTomo/CRMod elem.dat mesh file from either a file, or from stringIO
- plot_grid(**kwargs)[source]¶
Plot the mesh
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- Returns:
fig (matplotlib.Figure) – The Figure object
ax (matplotlib.Axes) – The axes object the mesh was plotted to
- plot_grid_to_ax(ax, **kwargs)[source]¶
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- reverse_node_order(element_type)[source]¶
Reverses the order of the nodes of all elements of a given type. This can be used to fix CRTomo/CRMod errors regarding the computation of the determinant during FE system compilation.
Use only if you know what you are doing!
- Parameters:
element_type (int) – Element type to change. Usually 3 or 4 (triangles or quads)
- class crtomo.eitMan(**kwargs)[source]¶
Bases:
objectManage sEIT data
- __init__(**kwargs)[source]¶
Data/frequency input: Note that only one of the parameters seitdir, crt_data_dir, seit_data will be used during initialization, in the stated priority.
Initialization diagram:
- Parameters:
seitdir (string, optional) – Load frequencies, grid, and data from an existing sEIT directory. Honors the shallow_import parameter.
shallow_import (bool, optional) – In combination with seitdir, only import the last iteration inversion results (faster).
crt_data_dir (string, optional) – if given, then try to load data from this directory. Expect a ‘frequencies.dat’ file and corresponding ‘volt_*.dat’ files.
only_frequency_ids (numpy.ndarray|None) – Load only the frequencies associated with these indices (zero-based). Only works with crt_data_dir.
seit_data (dict) – A dictionary with frequencies as keys, and numpy arrays as items. Each array must have 6 columns: a,b,m,n,magnitude[Ohm],phase[mrad]
frequencies (numpy.ndarray, optional) – frequencies that we work with
grid (crtomo.crt_grid.crt_grid, optional) – A grid instance
elem_file (string, optional) – Path to elem file
elec_file (string, optional) – Path to elec file
decouplings_file (str, optional) – Path to decoupling file: For not will be copied to the exe/ subdir of each tomodir written
crmod_cfg (?, optional)
crtomo_cfg (?, optional)
parman (?, optional)
- _init_frequencies(frequencies, configs_abmn=None)[source]¶
Initialize the tdMan instances associated with each frequency
Note that existing tds will not be deleted/overwritten
- Parameters:
frequencies (Nx1 numpy.ndarray) – Frequencies in ascending order
configs_abmn (None|numpy.ndarray (Mx4)) – Measurement configurations to provide to the generated tdMan instances
- add_homogeneous_model(magnitude, phase=0, frequency=None)[source]¶
Add homogeneous models to one or all tomodirs. Register those as forward models
- add_to_configs(configs)[source]¶
Add configurations to all tomodirs
- Parameters:
configs (
numpy.ndarray) – Nx4 numpy array with abmn configurations
- assign_sip_signatures_using_mask(mask_raw, lookup_table)[source]¶
- Parameters:
lookup_table (dict)
spectrum (sip_response) – SIP spectrum to use for parameterization
- extract_all_spectra(label)[source]¶
Extract all SIP spectra, and return frequencies and a numpy array
- Parameters:
label (str) – the label (data type) to extract. This corresponds to a key in eitMan.assignments. Possible values are rmag, rpha, cre, cim
- extract_points(label, points)[source]¶
Extract data points (i.e., SIP spectra) for one or more points.
- Parameters:
label (str) – the label (data type) to extract. This corresponds to a key in eitMan.assignments. Possible values are rmag, rpha, cre, cim
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
df_all – A dataframe with the extracted data
- Return type:
- get_measurement_responses()[source]¶
Return a dictionary of sip_responses for the modeled SIP spectra
Note that this function does NOT check that each frequency contains the same configurations!
- Returns:
responses – Dictionary with configurations as keys
- Return type:
- load_data_crt_files(data_dict, **kwargs)[source]¶
Load sEIT data from .ctr files (volt.dat files readable by CRTomo, produced by CRMod)
- Parameters:
data_dict (dict) – Data files that are imported. See example down below
Examples
>>> import glob data_files = {} data_files['frequencies'] = 'data/frequencies.dat' files = sorted(glob.glob('data/volt_*.crt')) data_files['crt'] = files
- load_inversion_results(sipdir, shallow_import=True)[source]¶
Given an sEIT inversion directory, load inversion results and store the corresponding parameter ids in self.assignments
Note that all previous data stored in this instance of the eitManager will be overwritten, if required!
- Parameters:
sipdir (string) – path to a CRTomo sip-invdir (i.e., a full sEIT inversion directory)
shallow_import (bool, optional) – If set to True, then only import the last inversion result, nothing else.
- measurements(**kwargs)[source]¶
Return modeled measurements. If not already done, call CRMod for each frequency to actually compute the forward response.
- Returns:
m_all –
dimension: frequency
dimension: config-number
dimension: 2: magnitude and phase (resistivity)
- Return type:
- model(**kwargs)[source]¶
Run the forward modeling for all frequencies.
Use
crtomo.eitManager.eitMan.measurements()to retrieve the resulting synthetic measurement spectra.- Parameters:
**kwargs (dict, optional) – All kwargs are directly provide to the underlying
crtomo.tdManager.tdMan.model()function calls.
- plot_forward_models(maglim=None, phalim=None, **kwargs)[source]¶
Create plots of the forward models
- Returns:
return_dict – Dictionary containing the figure and axes objects of the magnitude plots
- Return type:
- plot_result_spectrum(filename, nr)[source]¶
Plot a given data and inversion response spectrum to file.
WARNING: At this point does not take into account missing configurations for certain frequencies…
- register_forward_model(frequency, mag_model, phase_model)[source]¶
Register a magnitude parameter set and optionally a phase parameter set as a forward model for a given frequency.
- Parameters:
frequency (float) – Frequency. Must match the frequencies in eitMan.tds
mag_model (numpy.ndarray) – The magnitude model (linear scale)
phase_model (numpy.ndarray) – The phase model [mrad]
- save_measurements_to_directory(directory)[source]¶
Store the measurements (either from a previous import, or from forward modeling, in one directory in the CRTomo ‘crt’-style.
Frequencies are stored in a file frequencies.dat.
For each frequency, data is stored in a file volt_%.2i.dat in the CRTomo format.
- save_to_eitdir(directory)[source]¶
Save the eit data into a eit/sip directory structure
- Parameters:
directory (string|path) – output directory
- set_area_to_single_colecole(xmin, xmax, zmin, zmax, ccpars)[source]¶
Parameterize a rectangular area of the forward model using a single-term Cole-Cole response.
- class crtomo.mesh(elem_file=None, elec_file=None, empty=False)[source]¶
Bases:
crt_grid- Wm()¶
Return the smoothing regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.48ff
\[\begin{split}\Psi_m = ||\underline{\underline{W}}_m \underline{m}||^2_2\\ = \sum_{j=1}^M \sum_{i=nb(j)} \alpha_{r_ij} |\frac{m_j - m_i}{\Delta c_{ij}|^2 \Delta b_{ij} \Delta c_{ij}\end{split}\]Note that the anisotropic regularization parameter \(\alpha\) is not implemented yet.
i and j are swapped in the implementation.
See Fig 3.5 in the thesis.
- Wm_mgs(m, beta)¶
Return the MGS regularization matrix Wm of the grid
See PhD Thesis Roland Blaschek, eq. 3.50
Note that alpha and f are set 1
- Parameters:
m (numpy.ndarray) – model parameters to compute MGS matrix for
beta (float) – MGS beta parameter
- Returns:
Wm – Sparse MGS matrix
- Return type:
- __init__(elem_file=None, elec_file=None, empty=False)¶
- static _get_area_polygon(points_x, points_z)¶
Return the area of a polygon. The node coordinates must be ordered, but the direction does not matter - we return the abs-value
>>> points_x = [4, 4, 8, 8, -4, -4] >>> points_z = [6, -4, -4, -8, -8, 6] >>> self._get_area_polygon(points_x, points_z) ... 128
- _prepare_grids()¶
depending on the type of grid (rectangular or triangle), prepare grids or triangle lists
- TODO: We want some nice way of not needing to know in the future if we
loaded triangles or quadratic elements.
- _read_elem_elements(fid)¶
Read all FE elements from the file stream. Elements are stored in the self.element_data dict. The keys refer to the element types:
3: Triangular grid (three nodes)
8: Quadrangular grid (four nodes)
11: Mixed boundary element
12: Neumann (no-flow) boundary element
- _read_elem_header(fid)¶
- _read_elem_neighbors(fid)¶
Read the boundary-element-neighbors from the end of the file
- _read_elem_nodes(fid)¶
Read the nodes from an opened elem.dat file. Correct for CutMcK transformations.
We store three typed of nodes in the dict ‘nodes’:
“raw” : as read from the elem.dat file
“presort” : pre-sorted so we can directly read node numbers from a elec.dat file and use them as indices.
- “sorted”completely sorted as in the original grid (before any
CutMcK)
For completeness, we also store the following keys:
“cutmck_index” : Array containing the indices in “presort” to obtain the “sorted” values: nodes[‘sorted’] = nodes[‘presort’] [nodes[‘cutmck_index’], :]
“rev_cutmck_index” : argsort(cutmck_index)
- _write_elem_header(fid)¶
- _write_elements(fid)¶
- _write_neighbors(fid)¶
- _write_nodes(fid)¶
- add_grid_data(data)¶
Return id
- analyze_internal_angles(return_plot=False)¶
Analyze the internal angles of the grid. Angles shouldn’t be too small because this can cause problems/uncertainties in the Finite-Element solution of the forward problem. This function prints the min/max values, as well as quantiles, to the command line, and can also produce a histogram plot of the angles.
- Parameters:
return_plot (bool) – if true, return (fig, ax) objects of the histogram plot
- Returns:
fig (matplotlib.figure) – figure object
ax (matplotlib.axes) – axes object
Examples
>>> import crtomo.grid as CRGrid grid = CRGrid.crt_grid() grid.load_elem_file('elem.dat') fig, ax = grid.analyze_internal_angles(Angles) This grid was sorted using CutMcK. The nodes were resorted! Triangular grid found Minimal angle: 22.156368696965796 degrees Maximal angle: 134.99337326279496 degrees Angle percentile 10%: 51.22 degrees Angle percentile 20%: 55.59 degrees Angle percentile 30%: 58.26 degrees Angle percentile 40%: 59.49 degrees Angle percentile 50%: 59.95 degrees Angle percentile 60%: 60.25 degrees Angle percentile 70%: 61.16 degrees Angle percentile 80%: 63.44 degrees Angle percentile 90%: 68.72 degrees generating plot... >>> # save to file with fig.savefig('element_angles.png', dpi=300)
- calculate_dimensions()¶
For a regular grid, calculate the element and node dimensions
- static create_surface_grid(nr_electrodes=None, spacing=None, electrodes_x=None, electrodes_z=None, depth=None, left=None, right=None, char_lengths=None, lines=None, internal_lines=None, debug=False, workdir=None, force_neumann_only=False)¶
This is a simple wrapper for cr_trig_create to create simple surface grids.
Electrode and boundary positions are rounded to the third digit.
- Parameters:
nr_electrodes (int, optional) – the number of surface electrodes
spacing (float, optional) – the spacing between electrodes, usually in [m], required if nr of electrodes is given
electrodes_x (array, optional) – x-electrode positions can be provided here, e.g., for non-equidistant electrode distances
electrodes_z (array, optional) – z-electrode positions can be provided here, e.g., for non-equidistant electrode distances. Only useful in combination with electrodes_x
depth (float, optional) – the depth of the grid, relative to the minimum z-value. If not given, this is computed as half the maximum distance between electrodes
left (float, optional) – the space allocated left of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
right (float, optional) – the space allocated right of the first electrode. If not given, compute as a fourth of the maximum inter-electrode distance
char_lengths (float|list of 4 floats, optional) – characteristic lengths, as used by cr_trig_create
lines (list of floats, optional) – at the given depths, add horizontal lines in the grid. Note that all positive values will be multiplied by -1!
internal_lines (list of 4-tuple) – extra lines to add to the grid. Important: These lines must NOT touch the outer edge of the grid (this is not supported by this function, but can be accomplished by manually building the grid)
debug (bool, optional) – default: False. If true, don’t hide the output of cr_trig_create
workdir (string, optional) – if set, use this directory to create the grid. Don’t delete files afterwards.
force_neumann_only (bool, optional) – sometimes we want to use only Neumann boundary conditions. Setting this switch to True will force all boundaries to Neumann. Use only if you know what you are doing! default: False
- Returns:
grid – the generated grid
- Return type:
crtomo.grid.crt_gridinstance
Examples
>>> from crtomo.grid import crt_grid >>> grid = crt_grid.create_surface_grid(40, spacing=0.25, depth=5, ... left=2, right=2, char_lengths=[0.1, 0.5, 0.1, 0.5], ... lines=[0.4, 0.8], debug=False, workdir=None) >>> import pylab as plt >>> fig, ax = plt.subplots() >>> grid.plot_grid_to_ax(ax)
- determine_path_along_nodes(start_coordinate, end_coordinate)¶
- class element¶
Bases:
object- center()¶
- length_line()¶
- outer_normal_line()¶
- vector_line()¶
Return the vector of the boundary element
- vector_norm_line()¶
- property element_neighbors¶
- property element_neighbors_old¶
Return a list with element numbers (zero indexed) of neighboring elements. Note that the elements are not sorted. No spacial orientation can be inferred from the order of neighbors.
WARNING: This function is slow due to a nested loop. This would be a good starting point for further optimizations.
In order to speed things up, we could search using the raw data, i.e., with CutMcK enabled sorting, and then restrict the loops to 2x the bandwidth (before - after).
While not being returned, this function also sets the variable self.element_neighbors_edges, in which the common nodes with each neighbor are stored.
- Returns:
neighbors – a list (length equal to nr of elements) with neighboring elements
- Return type:
Examples
- find_nearest_node(coords, return_node_coords=True)¶
- get_distance_matrix()¶
- get_electrode_node(electrode)¶
For a given electrode (e.g. from a config.dat file), return the true node number as in self.nodes[‘sorted’]
- get_electrode_positions()¶
Return the electrode positions in an numpy.ndarray
- Returns:
positions – Nx2 array, where N is the number of electrodes. The first column contains x positions, the second z positions.
- Return type:
- get_element_areas()¶
return the areas of the elements
note that this formula is generic, see CRMod code for a simpler one
- Returns:
areas
- Return type:
- get_element_centroids()¶
return the central points of all elements
- Returns:
centroids – Nx2 array x/z coordinates for all (N) elements
- Return type:
- get_element_indices_along_line(p0, p1, N)¶
- get_element_indices_within_rectangle(xmin, xmax, zmin, zmax)¶
Return the indices of all elements whose center is located within the rectangle defined by the parameters.
The indices can then be used, e.g., to select values from inversion results.
- Parameters:
- Returns:
indices – Array with indices (zero-indexed)
- Return type:
np.array
- get_internal_angles()¶
Compute all internal angles of the grid
- Returns:
NxK array with N the number of elements, and K the number of nodes, filled with the internal angles in degrees
- Return type:
- get_min_max_electrode_distances()¶
Return the minimal and the maximal electrode distance of the grid
- Returns:
amin (float) – minimal electrode distance
amax (float) – maximal electrode distance
- get_minmax()¶
Return min/max x/z coordinates of grid
- Returns:
x ([float, float]) – min, max values of grid dimensions in x direction (sideways)
z ([float, float]) – min, max values of grid dimensions in z direction (downwards)
- get_node_tree()¶
- get_polygon_from_file(filename)¶
- static interpolate_grid(ingrid, outgrid, data, method='nearest')¶
Function for interpolating data from one grid to another, using the cell midpoints as datapoint locations. Standard method for interpolating is set to nearest-neighbour.
- Parameters:
ingrid (
crtomo.grid.crt_gridinstance) – CRT grid that matches the input dataoutgrid (
crtomo.grid.crt_gridinstance) – CRT grid to interpolate todata (pandas.dataframe) – input data that matches the input grid (in pandas dataframe format)
method (interpolation method, optional) – Standard interpolation method is nearest-neighbour (‘nearest’). Other possible methods are ‘linear’ and ‘cubic’.
- Returns:
interpolated data – returned dataframe with interpolated data
- Return type:
pandas.dataframe
- load_elec_file(elec_file)¶
- load_elem_file(elem_file)¶
Load a CRTomo/CRMod elem.dat mesh file from either a file, or from stringIO
- load_grid(elem_file, elec_file)¶
Load elem.dat and elec.dat
- plot_grid(**kwargs)¶
Plot the mesh
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- Returns:
fig (matplotlib.Figure) – The Figure object
ax (matplotlib.Axes) – The axes object the mesh was plotted to
- plot_grid_to_ax(ax, **kwargs)¶
- Parameters:
plot_electrode_numbers (bool, optional) – Plot electrode numbers in the grid, default: False
- reverse_node_order(element_type)¶
Reverses the order of the nodes of all elements of a given type. This can be used to fix CRTomo/CRMod errors regarding the computation of the determinant during FE system compilation.
Use only if you know what you are doing!
- Parameters:
element_type (int) – Element type to change. Usually 3 or 4 (triangles or quads)
- save_elec_file(filename)¶
- save_elem_elec_files(file_elem='elem.dat', file_elec='elec.dat')¶
- save_elem_file(output)¶
Save elem.dat to file. The grid is saved as read in, i.e., with or without applied cutmck. If you want to change node coordinates, use self.nodes[‘raw’]
- Parameters:
filename (string) – output filename
- test_plot()¶
- class crtomo.noise_model(seed, mag_rel=0, mag_abs=0, pha_a1=0, pha_b1=0, pha_rel=0, pha_abs=0)[source]¶
Bases:
objectnoise model as contained in the crt.noisemod file
Notes
line: 1 # Ensemble seed
line: 0.300 # Relative error resistance A (noise) [%] dR=AR+B
line: 0.100E-01 # Absolute errior resistance B (noise) [Ohm m]
line: 0.00 # Phase error parameter A1 [mRad/Ohm/m] dp=A1*R^B1+A2*p+p0
line: 0.00 # Phase error parameter B1 (noise) []
line: 0.00 # Relative phase error A2 (noise) [%]
line: 0.00 # Absolute phase error p0 (noise) [mRad]
- crtomo.pltMan¶
alias of
plotManager
- class crtomo.tdMan(**kwargs)[source]¶
Bases:
objectManage tomodirs
- __init__(**kwargs)[source]¶
The following initialization permutations are possible:
initialize empty
- initialize from an existing tomodir
tomodir: [tomodir path]
- supply one or more of the building blocks of a tomodir
grid: crtomo.grid.crt_grid instance
crmod_cfg: crtomo.cfg.crmod_cfg instance
crtomo_cfg: crmod.cfg.crtomo_cfg instance
http://www2.geo.uni-bonn.de/~mweigand/dashboard/content/crtomo_doc/crtomo/files.html#inv
- Keyword Arguments:
grid (crtomo.grid.crt_grid) – A fully initialized grid object
tempdir (string|None, optional) – if set, use this directory to create all temporary directories in. For example, settings this to /dev/shm can result in faster generation of large Jacobians
volt_file (None|str|numpy.ndarray) – if this is None, assume we didn’t get any measurement data. If this is a str, assume it to be the filename to a CRTomo volt.dat file. If it is a numpy array, assume 6 columns: a,b,m,n,rmag,rpha
volt_data (synonym for volt_file parameter) – see description for volt_file
- _initialize_components(kwargs)[source]¶
initialize the various components using the supplied **kwargs
- Parameters:
kwargs (dict) – kwargs dict as received by __init__()
- _invert(tempdir, catch_output=True, **kwargs)[source]¶
Internal function than runs an inversion using CRTomo.
- Parameters:
- Returns:
success (bool) – False if an error was detected
error_msg (str) – Error message. None if not error was encountered.
output (str) – Output of the actual CRTomo call
- static _read_eps_ctr(tomodir)[source]¶
Parse a CRTomo eps.ctr file.
TODO: change parameters to only provide eps.ctr file
- Parameters:
tomodir (string) – Path to directory path
- _read_inv_ctr(tomodir)[source]¶
Read in selected results of the inv.ctr file
- Parameters:
tomodir (string) – directory path to a tomodir
- Returns:
inv_ctr – structure containing inv.ctr data
- Return type:
?
- _read_inversion_fwd_responses(tomodir)[source]¶
Import the forward responses for all iterations of a given inversion
- Parameters:
tomodir (str) – Path to tomodir
- _read_inversion_results(tomodir)[source]¶
Import resistivity magnitude/phase and real/imaginary part of conductivity for all iterations
- Parameters:
tomodir (str) – Path to tomodir
- _read_l1_coverage(tomodir)[source]¶
Read in the L1 data-weighted coverage (or cumulated sensitivity) of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_l2_coverage(tomodir)[source]¶
Read in the L2 data-weighted coverage (or cumulated sensitivity) of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_modeling_results(mod_directory, silent=False)[source]¶
Read modeling results from a given mod/ directory. Possible values to read in are:
voltages
potentials
sensitivities
- _read_resm_m(tomodir)[source]¶
Read in the diagonal entries of the resolution matrix of an inversion
- Parameters:
tomodir (str) – directory path to a tomodir
- _read_sensitivities(sens_dir)[source]¶
import sensitivities from a directory
Note
check that signs are correct in case CRMod switches potential electrodes
- add_homogeneous_model(magnitude, phase=0)[source]¶
Add a homogeneous resistivity model to the tomodir. This is useful for synthetic measurements.
- Parameters:
- Returns:
pid_mag (int) – ID value of the parameter set of the magnitude model
pid_pha (int) – ID value of the parameter set of the phase model
Note that the parameter sets are automatically registered as the
forward models for magnitude and phase values.
- check_measurements_against_sensitivities(magnitude, phase=0, return_plot=False)[source]¶
Check for all configurations if the sensitivities add up to a given homogeneous model
- Parameters:
- Returns:
results (Nx6 numpy.ndarray) – Results of the analysis.
magnitude measurement [Ohm]
sum of sensitivities [Volt]
relative deviation of sensitivity-sum from measurement [in percent]
fig (matplotlib.figure, optional) – figure object. Only returned of return_plot=True
axes (list) – list of axes corresponding to the figure
Examples
>>> #!/usr/bin/python import crtomo.tdManager as CRtdMan tdm = CRtdMan.tdMan( elem_file='grid/elem.dat', elec_file='grid/elec.dat', config_file='config/config.dat', ) results, fig, axes = tdm.check_measurements_against_sensitivities( magnitude=100, phase=-10, return_plot=True ) fig.savefig('sensitivity_comparison.png', dpi=300)
- copy()[source]¶
Provide a copy of yourself. Do not copy modeling or inversion results, but copy everything that can be used for modeling or inversion
- extract_along_line(pid, xy0, xy1, N=10)¶
Extract parameter values along a given line.
- Parameters:
- Returns:
values – data values for extracted data points. First column: x-coordinates, second column: z-coordinates, third column: extracted values
- Return type:
numpy.ndarray (n x 3)
- extract_points(pid, points)¶
Extract values at certain points in the grid from a given parameter set. Cells are selected by interpolating the centroids of the cells towards the line using a “nearest” scheme.
Note that data is only returned for the points provided. If you want to extract multiple data points along a line, defined by start and end point, use the extract_along_line function.
- Parameters:
pid (int) – The parameter id to extract values from
points (Nx2 numpy.ndarray) – (x, y) pairs
- Returns:
values – data values for extracted data points
- Return type:
numpy.ndarray (n x 1)
- get_fwd_reda_container()[source]¶
Return a REDA container, either reda.ERT, or reda.CR, with modeled data
- Returns:
container
- Return type:
reda.ERT|reda.CR|None
- get_potential(config_nr)[source]¶
Return potential data for a given measurement configuration.
- Parameters:
config_nr (int) – Number of the configurations. Starts at 0
- Returns:
pot_data – First array: magnitude potentials, second array: phase potentials
- Return type:
list with two numpy.ndarrays
- get_sensitivity(config_nr)[source]¶
return a sensitivity, as well as corresponding metadata, for a given measurement configuration. Indices start at zero.
- inv_get_last_pid(parameter)[source]¶
Return the pid of the parameter set corresponding to the final inversion results of a given parameter. Return None if the parameter type does not exist, or no inversion result was registered.
- inv_last_cim_parset()[source]¶
- Return the imaginary part of the complex resistivity of the last
inversion iteration.None if no inversion data exists.
- Returns:
inv_last_cim
- Return type:
numpy.ndarray|None
- inv_last_cre_parset()[source]¶
- Return the real part of the complex resistivity of the last
inversion iteration. None if no inversion data exists.
- Returns:
inv_last_cre
- Return type:
numpy.ndarray|None
- inv_last_rmag_parset()[source]¶
Return the resistivity magnitude of the last iteration. None if no inversion data exists.
Example
>>> import crtomo ... tdm = crtomo.tdMan('tomodir/') ... tdm.inv_last_rmag_parset()
- Returns:
inv_last_rmag
- Return type:
numpy.ndarray|None
- inv_last_rpha_parset()[source]¶
- Return the phase magnitude of the last inversion iteration.
None if no inversion data exists.
- Returns:
inv_last_rpha
- Return type:
numpy.ndarray|None
- invert(output_directory=None, catch_output=True, **kwargs)[source]¶
Invert this instance, and import the result files
No directories/files will be overwritten. Raise an IOError if the output directory exists.
- Parameters:
output_directory (string, optional) – use this directory as output directory for the generated tomodir. If None, then a temporary directory will be used that is deleted after data import.
catch_output (bool, optional) – if True, do not show CRTomo output. Default:True
cores (int, optional) – how many cores to use for CRTomo
**kwargs (Are supplied to
crtomo.tdMan._invert())
- Returns:
return_code – Return 0 if the inversion completed successfully. Return 1 if no measurements are present.
- Return type:
- load_rho_file(filename)[source]¶
Load a forward model from a rho.dat file
- Parameters:
filename (string) – filename to rho.dat file
- Returns:
pid_mag (int) – parameter id for the magnitude model
pid_pha (int) – parameter id for the phase model
- measurements(silent=False)[source]¶
Return the measurements associated with this instance.
If measurements are not present, check if we can model, and then run CRMod to load the measurements.
- Parameters:
silent (bool, optional) – If False, suppress CRMod output
- model(voltages=True, sensitivities=False, potentials=False, output_directory=None, silent=False)[source]¶
Forward model the tomodir and read in the results
Note that this function will always do the full modeling. No caching involved.
Please use .measurements to accessed cached measurements.
- Parameters:
voltages (bool, optional) – if True, compute voltages for registered quadrupoles. Default: True
sensitivities (bool, optional) – if True, compute sensitivities for registered quadrupoles. Default: False
potentials (bool, optional) – if True, compute potential fields for all current injections. Default: False TODO: check if implemented in the Python wrapper
output_directory (str|None, optional) – if this is a string, treat it as an output directory in which the tomodir used for the modeling is saved. Will raise an exception if the output directory already exists. Default: None
silent (bool, optional) – if True, suppress most of the output. Default: False
- Return type:
None
- plot_decouplings_to_ax(ax, plot_transistions=True)[source]¶
Visualize decouplings. Usually you may want to plot the mesh to this axis first
- Parameters:
ax (matplotlib.Axes) – Axis to plot to
plot_transistions (bool (True)) – If True, then also visualize the decoupled transitions
- plot_eps_data(filename=None)[source]¶
Plot data residuals and data error weighting
- Parameters:
filename (string|None) – if not None, then save plot to this file
- Returns:
figure – if filename is None, then return the generated figure
- Return type:
matplotlib.Figure|None
- plot_eps_data_hist(filename=None)[source]¶
Plot histograms of data residuals and data error weighting
- TODO :
add percentage of data below/above the RMS value
- Parameters:
filename (string|None) – if not None, then save plot to this file
- Returns:
figure – if filename is None, then return the generated figure
- Return type:
matplotlib.Figure|None
- plot_forward_models(mag_only=False, separate_figures=False, **kwargs)[source]¶
Plot the forward models (requires the forward models to be loaded)
- plot_inversion_evolution(filename=None)[source]¶
Plot the evolution of inversion properties (lambda, RMS, …) :param filename: if not None, save plot to this file :type filename: string|None
- Returns:
fig – if filename is None, return the figure object
- Return type:
matplotlib.Figure|None
- plot_inversion_result_rmag(figsize=None, log10=False, overlay_coverage=False, **kwargs)[source]¶
Plot the final inversion results, magnitude-results only
- Parameters:
figsize ((float, float)|None) – Figure size of the matplotlib figure in inches.
log10 (bool, default: False) – Shortcut to force a log10 conversion of the resistivity data
overlay_coverage (bool, default: False) – If True, use the cumulated coverage to adjust the transparency of the plot
**kwargs (dict) – will be propagated into self.plot.plot_elements_to_ax
- Returns:
fig (matplotlib.Figure) – The created figure
ax (matplotlib.Axes) – Plot axes
- plot_pseudo_locs(spacing=1.0)[source]¶
Plot pseudo-locations of measurement configurations. This function does not take into account real electrode locations. However, for surface configurations a ‘spacing’ parameter can be provided
- Parameters:
spacing (float, default=1.0) – Electrode spacing
- plot_sensitivity(config_nr=None, sens_data=None, mag_only=False, absv=False, **kwargs)[source]¶
Create a nice looking plot of the sensitivity distribution for the given configuration nr. Configs start at 1!
- Parameters:
config_nr (int, optional) – The configuration number (starting with 0) to compute the sensitivity for.
sens_data (Nx2 numpy.ndarray, optional) – If provided, use this data as sensitivity data (do not compute anything)
mag_only (bool, optional) – Plot only the magnitude sensitivities
absv (bool, optional) – If true, plot absolute values of sensitivity
- Returns:
fig
ax
Examples
(
Source code,png,hires.png,pdf)
- read_decouplings_file(filename)[source]¶
Import decoupling data for the inversion. This is usally a file called decouplings.dat in the exe/ directory of a tomodir, but we can also read from an BytesIO object.
Do nothing if the file does not exist
Overwrite any existing decouplings
- read_inversion_results(tomodir)[source]¶
Import inversion results from a tomodir into this instance
Warning
This function is not finished yet and does not import ALL crtomo information yet.
- Parameters:
tomodir (str) – Path to tomodir
- read_voltages(voltage_file)[source]¶
Import voltages from a volt.dat file
- Parameters:
voltage_file (str) – Path to volt.dat file
- register_data_errors(mid_rmag_error, mid_rpha_error=None, norm_mag=1, norm_pha=1)[source]¶
Register individual data errors.
The normalization factors are used as follows:
mag_error -> mag_error / norm_mag ** 2 pha_error -> pha_error / norm_pha ** 2
That means that you need to decrease the values in order to increase individual errors.
- Parameters:
mid_mag_error (int) – ID to the corresponding measurement in tdMan.configs holding the magnitude errors (linear)
mid_pha_error (int) – ID to the corresponding measurement in tdMan.configs holding the phase errors (linear)
norm_mag (float) – Magnitude errors can be normalized by the square of this value.
norm_pha (float) – Phase errors can be normalized by the square of this value.
- register_forward_model(pid_mag, pid_pha)[source]¶
Register parameter sets as the forward models for magnitude and phase
- register_measurements(mag, pha=None)[source]¶
Register measurements as magnitude/phase measurements used for the inversion
- Parameters:
mag (int|numpy.ndarray) – magnitude measurement id for the corresponding measurement data in self.configs.measurements. If mag is a numpy.ndarray, assume mag to be the data itself an register it
pha (int, optional) – phase measurement id for the corresponding measurement data in self.configs.measurements. If not present, a new measurement set will be added with zeros only.
- reset_data()[source]¶
Attempts to reset (delete) all inversion data currently stored in the tdMan instance. This is mostly attempted for the impedance data (magnitudes, phases, conductivity real and imaginary parts), but could be extended to other data (this is currently not done due to complexity and missing demand). Forward models are also deleted.
- save_measurements(filename)[source]¶
Save measurements in a file. Use the CRTomo format.
Do not save anything if no measurements are present in this tdMan instance.
- Parameters:
filename (string) – path to output filename
- save_to_tarfile()[source]¶
Save the current modeling/inversion data into a tarfile. Return the file as an io.BytesIO object. The tar file is generated completely in memory - no files are written to the disc
- Returns:
tomodir_tar – Tomodir stored in tar file
- Return type:
- save_to_tomodir(directory, only_for_inversion=False)[source]¶
Save the tomodir instance to a directory structure.
At this point forward modeling results (voltages, potentials and sensitivities) will be saved.
Inversion results will, at this stage, not be saved (TODO!!!).
- Parameters:
directory (str) – Output directory (tomodir). Will be created of it does not exists, but otherwise files we be overwritten in any existing directories
only_for_inversion (bool, optional) – If True, save only files required for an inversion (i.e., omit any forward modeling files and results not necessary). Default: False
Note
Test cases:
modeling only
inversion only
modeling and inversion
- show_parset(pid, **kwargs)[source]¶
Plot a given parameter set. Thin wrapper around
crtomo.plotManager.plotManager.plot_elements_to_ax().kwargs will be directly supplied to the plot function
- crtomo.td_is_finished(tomodir)[source]¶
Return the state of modeling and inversion for a given tomodir. The result does not take into account sensitivities or potentials, as optionally generated by CRMod.
- Parameters:
tomodir (string) – Directory to check
- Returns:
crmod_is_finished (bool) – True if a successful CRMod result is contained in the tomodir directory.
crtomo_is_finished (bool) – True if a successful CRTomo inversion results is contained in the tomodir directory.