Note
Go to the end to download the full example code.
A single-frequency inversion (CR container)¶
Full processing of one frequency of one timestep of the data from Weigand and Kemna 2017 (Biogeosciences).
This example uses the CR container to load single-frequency data from a CRTomo-style volt.dat file.
imports
import os
import numpy as np
import crtomo
import reda
import reda.utils.geometric_factors as geom_facs
from reda.utils.fix_sign_with_K import fix_sign_with_K
import reda.importers.eit_fzj as eit_fzj
define an output directory for all files
output_directory = 'output_single_freq_inversion_CR'
cr = reda.CR()
cr.import_crtomo_data('data/volt.dat')
# this is a container measurement, we need to compute geometric factors using
# numerical modeling
# Note that this only works if CRMod is available
settings = {
'rho': 100,
'elem': 'data/elem.dat',
'elec': 'data/elec.dat',
'sink_node': '6467',
'2D': True,
}
k = geom_facs.compute_K_numerical(cr.data, settings)
cr.data = geom_facs.apply_K(cr.data, k)
fix_sign_with_K(cr.data)
print(cr.data.iloc[0:5])
SETTINGS
{'rho': 100, 'elem': 'data/elem.dat', 'elec': 'data/elec.dat', 'sink_node': '6467', '2D': True}
2D modeling
a b m n ... k rho_a sigma_a rho_a_complex
0 1 2 21 20 ... 9061.232484 46.166980 0.021661 40.270914+ 22.575286j
1 1 27 30 28 ... 1.529047 1282.753055 0.000780 1282.753033- 0.235647j
2 1 27 31 29 ... 1.596560 1298.674366 0.000770 1298.674241- 0.569899j
3 1 27 32 28 ... 0.689674 1336.160025 0.000748 1336.159970- 0.383912j
4 1 27 32 30 ... 1.256348 1380.042077 0.000725 1380.041984- 0.505736j
[5 rows x 15 columns]
apply correction factors, as described in Weigand and Kemna, 2017 BG
corr_facs_nor = np.loadtxt('data/corr_fac_avg_nor.dat')
corr_facs_rec = np.loadtxt('data/corr_fac_avg_rec.dat')
corr_facs = np.vstack((corr_facs_nor, corr_facs_rec))
cr.data, cfacs = eit_fzj.apply_correction_factors(cr.data, corr_facs)
apply some filters import IPython IPython.embed()
cr.filter('r < 0')
cr.filter('rho_a < 15 or rho_a > 35')
cr.filter('rpha < - 40 or rpha > 3')
cr.filter('rphadiff < -5 or rphadiff > 5')
cr.filter('k > 400')
cr.filter('rho_a < 0')
cr.filter('a == 12 or b == 12 or m == 12 or n == 12')
cr.filter('a == 13 or b == 13 or m == 13 or n == 13')
# NOTE: this is also a single-frequency filtering,
cr.print_data_journal()
cr.print_log()
--- Data Journal Start ---
2026-01-12 10:27:37.015476
A filter was applied with query "r < 0". In total 15 records were removed
A filter was applied with query "rho_a < 15 or rho_a > 35". In total 394 records were removed
A filter was applied with query "rpha < - 40 or rpha > 3". In total 159 records were removed
A filter was applied with query "rphadiff < -5 or rphadiff > 5". In total 82 records were removed
A filter was applied with query "k > 400". In total 224 records were removed
A filter was applied with query "rho_a < 0". In total 0 records were removed
A filter was applied with query "a == 12 or b == 12 or m == 12 or n == 12". In total 36 records were removed
A filter was applied with query "a == 13 or b == 13 or m == 13 or n == 13". In total 33 records were removed
--- Data Journal End ---
2026-01-12 10:27:30,469 - reda.containers.BaseContainer - INFO - Data sized changed from 1925 to 1910
2026-01-12 10:27:31,636 - reda.containers.BaseContainer - INFO - Data sized changed from 1910 to 1516
2026-01-12 10:27:32,598 - reda.containers.BaseContainer - INFO - Data sized changed from 1516 to 1357
2026-01-12 10:27:33,498 - reda.containers.BaseContainer - INFO - Data sized changed from 1357 to 1275
2026-01-12 10:27:34,354 - reda.containers.BaseContainer - INFO - Data sized changed from 1275 to 1051
2026-01-12 10:27:35,041 - reda.containers.BaseContainer - INFO - Data sized changed from 1051 to 1051
2026-01-12 10:27:35,727 - reda.containers.BaseContainer - INFO - Data sized changed from 1051 to 1015
2026-01-12 10:27:36,384 - reda.containers.BaseContainer - INFO - Data sized changed from 1015 to 982
export to volt.dat file note that this is not required for the subsequent code
with reda.CreateEnterDirectory(output_directory):
cr.export_crtomo('volt.dat', 'nor')
alternatively: directly create a tdman object that represents a single-frequency inversion with CRTomo
grid = crtomo.crt_grid('data/elem.dat', 'data/elec.dat')
tdman = cr.export_to_crtomo_td_manager(grid, norrec='nor')
# set inversion settings
tdman.crtomo_cfg['robust_inv'] = 'F'
tdman.crtomo_cfg['mag_abs'] = 0.012
tdman.crtomo_cfg['mag_rel'] = 0.5
tdman.crtomo_cfg['hom_bg'] = 'T'
tdman.crtomo_cfg['d2_5'] = 0
tdman.crtomo_cfg['fic_sink'] = 'T'
tdman.crtomo_cfg['fic_sink_node'] = 6467
# run the inversion, use the given output directory to store the CRTomo
# directory structure for later use
# only invert if the output directory does not exists
outdir = '{}/tomodir_inversion'.format(output_directory)
if not os.path.isdir(outdir):
tdman.invert(
catch_output=False,
output_directory=outdir,
)
else:
tdman.read_inversion_results(outdir)
print('Statistics of last iteration:')
print(tdman.inv_stats.iloc[-1])
This grid was sorted using CutMcK. The nodes were resorted!
Rectangular grid found
Attempting inversion in directory: output_single_freq_inversion_CR/tomodir_inversion
Using binary: /usr/bin/CRTomo_dev
Calling CRTomo
Inversion attempt finished
Attempting to import the results
Reading inversion results
is robust False
Info: res_m.diag not found: output_single_freq_inversion_CR/tomodir_inversion/inv/res_m.diag
/home/runner/work/crtomo_tools/crtomo_tools/examples/02_simple_inversion
Info: ata.diag not found: output_single_freq_inversion_CR/tomodir_inversion/inv/ata.diag
/home/runner/work/crtomo_tools/crtomo_tools/examples/02_simple_inversion
Statistics of last iteration:
iteration 2
main_iteration 2
it_type DC/IP
type main
dataRMS 1.0
magRMS 1.0
phaRMS 0.0
lambda 476.2
roughness 0.4061
cgsteps 62.0
nrdata 477.0
steplength 0.0
stepsize 61.68
l1ratio NaN
Name: 11, dtype: object
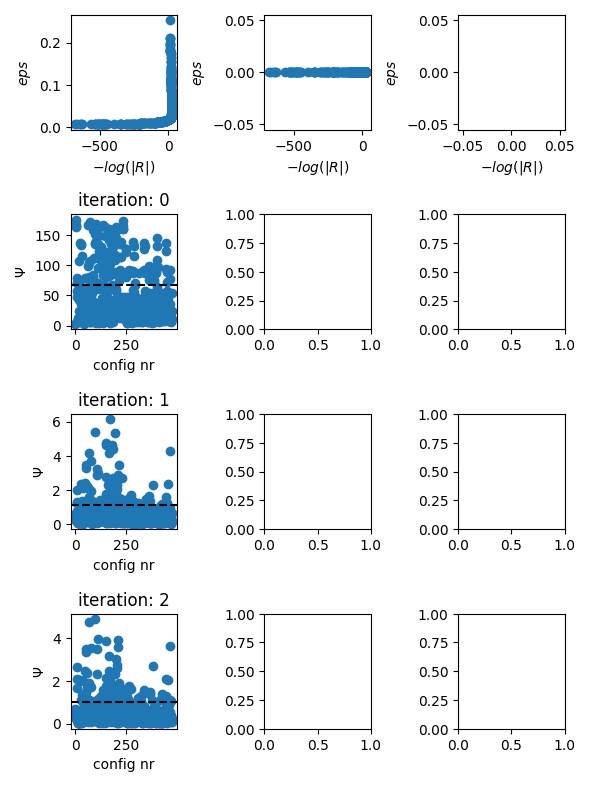
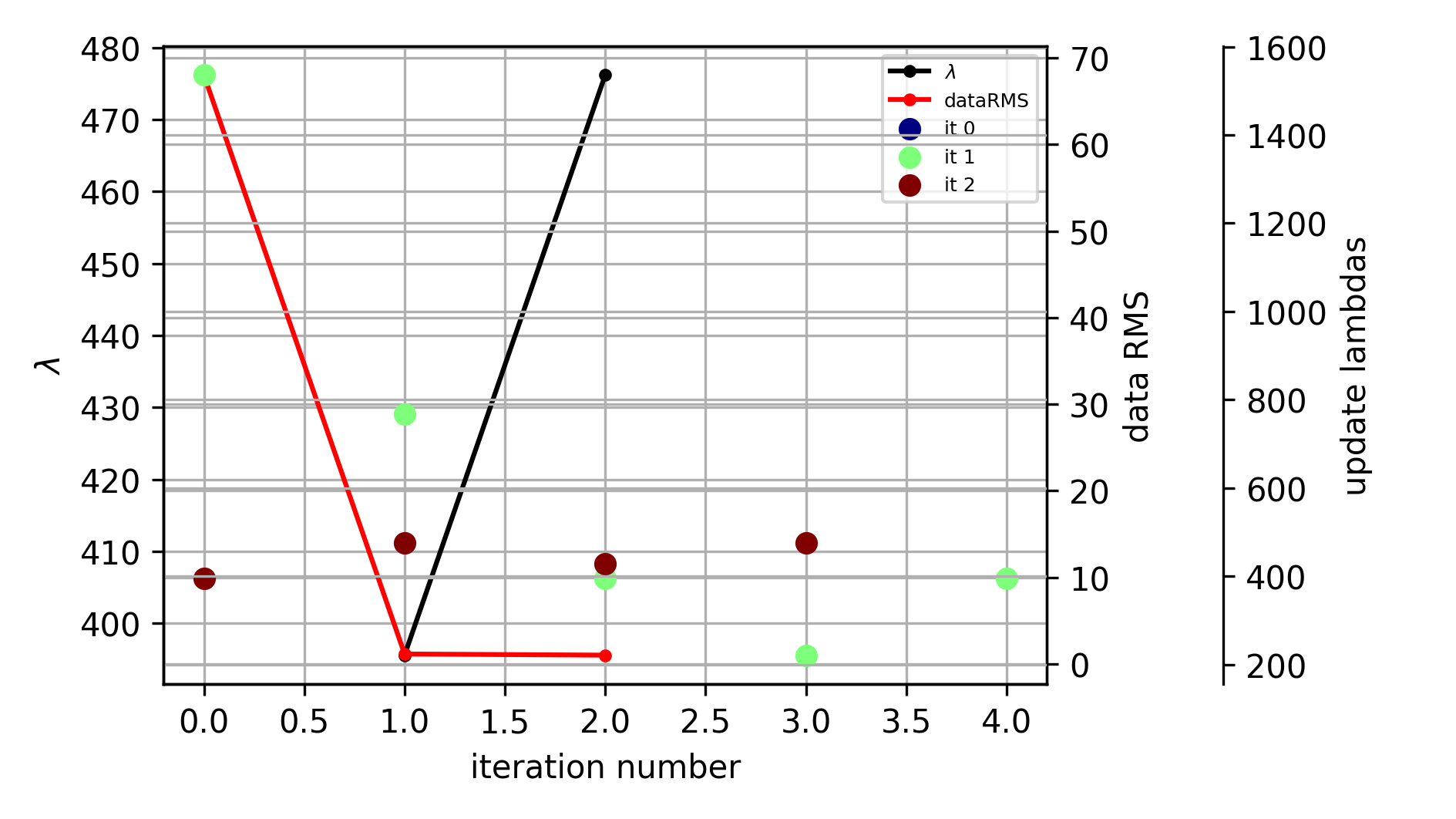
evolution of the inversion
fig = tdman.plot_inversion_evolution()
with reda.CreateEnterDirectory(output_directory):
fig.savefig('inversion_evolution.png')
# the statistics are stored in a data frame
print(tdman.inv_stats)
print(tdman.inv_stats.columns)
#############################
0
Empty DataFrame
Columns: [iteration, main_iteration, it_type, type, dataRMS, magRMS, phaRMS, lambda, roughness, cgsteps, nrdata, steplength, stepsize, l1ratio]
Index: []
#############################
1
iteration main_iteration it_type ... steplength stepsize l1ratio
1 1 1 DC/IP ... 1.0 7440.0 NaN
2 2 1 DC/IP ... 1.0 7370.0 NaN
3 3 1 DC/IP ... 1.0 7290.0 NaN
4 4 1 DC/IP ... 1.0 7200.0 NaN
5 5 1 DC/IP ... 0.5 3650.0 NaN
[5 rows x 14 columns]
#############################
2
iteration main_iteration it_type ... steplength stepsize l1ratio
7 0 2 DC/IP ... 1.0 61.0 NaN
8 1 2 DC/IP ... 1.0 61.7 NaN
9 2 2 DC/IP ... 1.0 61.3 NaN
10 3 2 DC/IP ... 0.5 30.8 NaN
[4 rows x 14 columns]
iteration main_iteration it_type ... steplength stepsize l1ratio
0 0 0 DC/IP ... NaN NaN NaN
1 1 1 DC/IP ... 1.0 7440.00 NaN
2 2 1 DC/IP ... 1.0 7370.00 NaN
3 3 1 DC/IP ... 1.0 7290.00 NaN
4 4 1 DC/IP ... 1.0 7200.00 NaN
5 5 1 DC/IP ... 0.5 3650.00 NaN
6 1 1 DC/IP ... 3.0 7293.00 NaN
7 0 2 DC/IP ... 1.0 61.00 NaN
8 1 2 DC/IP ... 1.0 61.70 NaN
9 2 2 DC/IP ... 1.0 61.30 NaN
10 3 2 DC/IP ... 0.5 30.80 NaN
11 2 2 DC/IP ... 0.0 61.68 NaN
[12 rows x 14 columns]
Index(['iteration', 'main_iteration', 'it_type', 'type', 'dataRMS', 'magRMS',
'phaRMS', 'lambda', 'roughness', 'cgsteps', 'nrdata', 'steplength',
'stepsize', 'l1ratio'],
dtype='object')
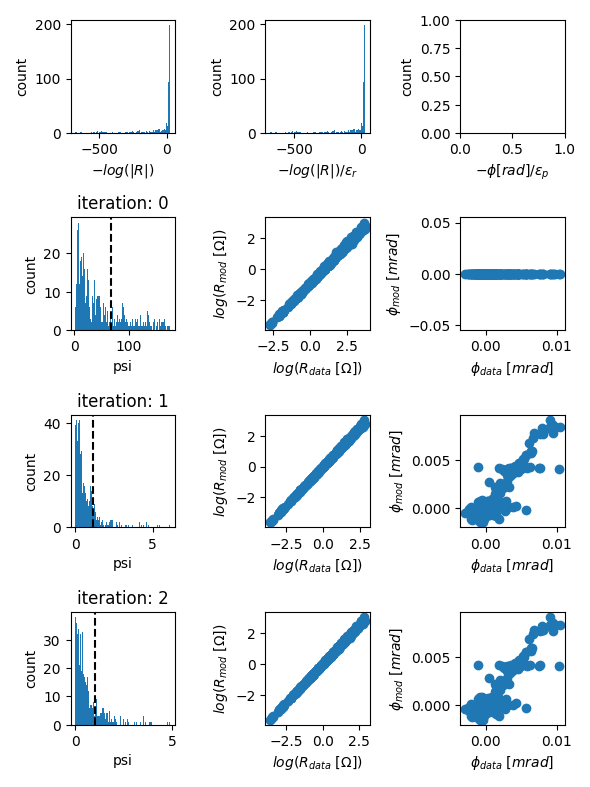
evolution of data misfits
fig = tdman.plot_eps_data()
fig = tdman.plot_eps_data_hist()
# eps data is found here:
tdman.eps_data
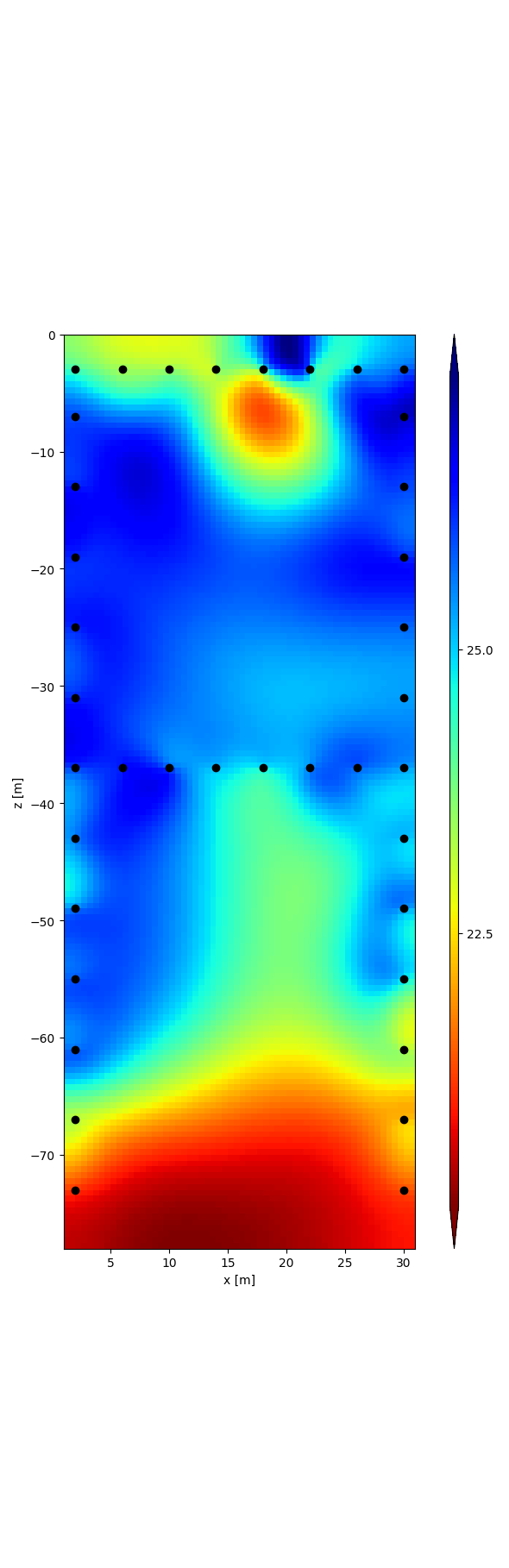
r = tdman.plot.plot_elements_to_ax(
tdman.a['inversion']['rmag'][-1],
plot_colorbar=True,
cmap_name='jet_r',
)
with reda.CreateEnterDirectory(output_directory):
r[0].savefig('rmag.png', bbox_inches='tight')
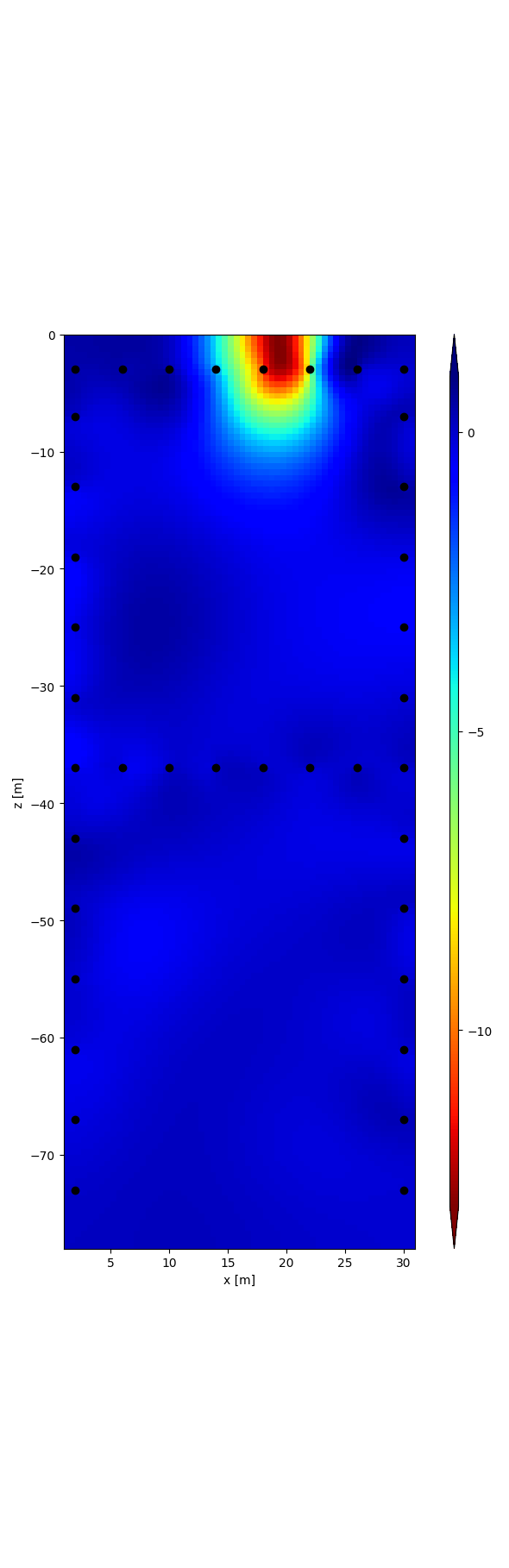
r = tdman.plot.plot_elements_to_ax(
tdman.a['inversion']['rpha'][-1],
plot_colorbar=True,
cmap_name='jet_r',
)
with reda.CreateEnterDirectory(output_directory):
r[0].savefig('rpha_last_it.png', bbox_inches='tight')
Total running time of the script: (2 minutes 26.309 seconds)