Note
Go to the end to download the full example code.
Using the tdManager to investigate inversion results¶
One of the purposes of the tdManager is to provide an easy interface to analyze
inversion results.
Inversion results can either be directly generated using the
crtomo.tdMan.invert() function, or by loading from an already-finished
inversion directory (the tomodir).
In this example we will explore the various functionalities of the tdManager.
we need the crtomo module to continue
import matplotlib.pylab as plt
import crtomo
# the pprint module is used just to prettify the output of this example, please
# ignore for further use
import pprint
Load previously-generated inversion results from an already-finished tomodir
tdm = crtomo.tdMan(tomodir='tomodir')
importing tomodir tomodir
This grid was sorted using CutMcK. The nodes were resorted!
Triangular grid found
importing tomodir results
loaded configs: (220, 2)
reading voltages
Reading inversion results
is robust True
The first order of business is to access the convergence behavior of the
inversion. This data is loaded into a pandas.DataFrame:
print(tdm.inv_stats)
# TODO Plot inv stats
iteration main_iteration it_type ... steplength stepsize l1ratio
0 0 0 DC/IP ... NaN NaN 1.14
1 1 1 DC/IP ... NaN 13200.00 NaN
2 2 1 DC/IP ... NaN 13600.00 NaN
3 3 1 DC/IP ... NaN 14200.00 NaN
4 4 1 DC/IP ... NaN 15300.00 NaN
.. ... ... ... ... ... ... ...
75 8 8 DC/IP ... NaN 48.90 NaN
76 9 8 DC/IP ... NaN 52.60 NaN
77 10 8 DC/IP ... NaN 56.60 NaN
78 11 8 DC/IP ... NaN 26.30 NaN
79 8 8 DC/IP ... 0.0 52.63 1.00
[80 rows x 14 columns]
Inversion results are stored as parameter sets in the ParMan instance
(crtomo.ParMan):
print(tdm.parman)
<crtomo.parManager.ParMan object at 0x7fc2677db760>
the ParMan stores multiple parameter sets (i.e., data sets which assign each cell of the FE-mesh a given value), identified by an integer index number, the parameter id, pid. The pids for the various parameters imported from the inversion results are stored in dictionary entries:
pprint.pprint(tdm.a['inversion'])
# We can see several entries in the dictionary: The resistivity magnitude
# (rmag), the phase value (rpha), the real part of the complex conductivity
# (cre), the imaginary part of the complex conductivity (cim) and an entry
# which combines cre and cim (cre_cim). For each inversion, the parameter sets
# have an own pid to make the results accessible.
{'cim': [19, 21, 23, 25, 27, 29, 31, 33, 35],
'cre': [18, 20, 22, 24, 26, 28, 30, 32, 34],
'cre_cim': [[18, 19],
[20, 21],
[22, 23],
[24, 25],
[26, 27],
[28, 29],
[30, 31],
[32, 33],
[34, 35]],
'fwd_response_rmag': [2, 5, 8, 11, 14, 17, 20, 23, 26],
'fwd_response_rpha': [3, 6, 9, 12, 15, 18, 21, 24, 27],
'fwd_response_wdfak': [4, 7, 10, 13, 16, 19, 22, 25, 28],
'l1_dw_log10_norm': 37,
'l2_dw_log10_norm': 38,
'resm': 36,
'rmag': [0, 1, 2, 3, 4, 5, 6, 7, 8],
'rpha': [9, 10, 11, 12, 13, 14, 15, 16, 17]}
To access a given parameter set, extract the pid from the dict and then access the data in the ParMan instance. If we would like to access the phase values for e.g. the second inversion run, we have to choose the index accordingly:
pid_rpha_2nd = tdm.a['inversion']['rpha'][1]
print('pid of the second phase inversion result: ', pid_rpha_2nd)
rpha_second_inversion = tdm.parman.parsets[pid_rpha_2nd]
print(
'Pixel values of the second phase inverion result: ',
rpha_second_inversion
)
pid of the second phase inversion result: 10
Pixel values of the second phase inverion result: [ -7.50958061 -14.9736643 -10.9377241 ... -2.00979733 -2.66891265
-12.5039339 ]
The last inversion result can be accessed using the index -1
rpha_last_inversion = tdm.parman.parsets[tdm.a['inversion']['rmag'][-1]]
Most of the time, it is of interest to get the final inversion result. This can be done in two seperate ways. Either by settings the index to [-1] or by using the built-in retriever function from crtomo-tools:
rmag_final_inversion = tdm.inv_last_rmag_parset()
# The function returns a numpy-array with the inversion results. A similar
# function exists to extract the final results for the phase value (rpha),
# the real part of the complex conductivity (cre) and the imaginary
# part of the complex conductivity (cim):
rpha_final_inversion = tdm.inv_last_rpha_parset()
cre_final_inversion = tdm.inv_last_cre_parset()
cim_final_inversion = tdm.inv_last_cim_parset()
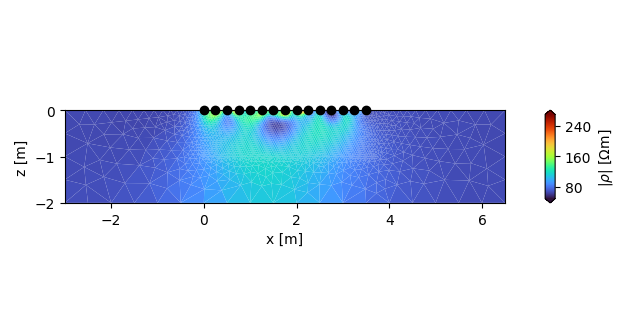
Plot the final magnitude result
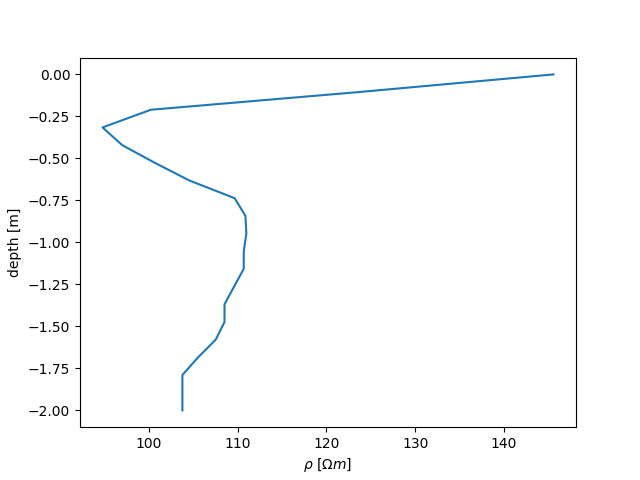
Now let us extract a vertical slice of the final magnitude result this is a Nx3 matrix: x, z, value columns
extracted_data = tdm.extract_along_line(
tdm.a['inversion']['rmag'][-1],
[2, 0],
[2, -2],
20
)
fig, ax = plt.subplots()
ax.plot(
extracted_data[:, 2],
extracted_data[:, 1],
)
ax.set_xlabel(r'$\rho~[\Omega m]$')
ax.set_ylabel('depth [m]')
# TODO: Extract from point, line, polygon, rectangle