Note
Go to the end to download the full example code.
Example for processing field data from Sycal¶
Note
This example is work in progress!!!
import matplotlib.pyplot as plt
import crtomo
import reda.exporters.crtomo as crto
import reda.utils.norrec as NR
import reda
Daten einlesen und berechnen¶
# reda container erstellen
ert = reda.ERT()
# normal und reciproke Daten einlesen
ert.import_syscal_bin('data_nor.bin')
ert.import_syscal_bin('data_rec.bin', reciprocals=48)
# K-Faktor berechnen
K = reda.utils.geometric_factors.compute_K_analytical(ert.data, spacing=2.5)
# Rho_a berechnen
reda.utils.geometric_factors.apply_K(ert.data, K)
# negative Widerstände bei negativen K umdrehen
reda.utils.fix_sign_with_K.fix_sign_with_K(ert.data)
# calculate diffeence between nor and rec measurement
ert.data = NR.assign_norrec_diffs(ert.data, ('r', 'rho_a'))
Histogramme plotten¶
fig = ert.histogram(('r', 'rdiff', 'rho_a', 'rho_adiff', 'Iab'))
plt.savefig('histogramms.png')
plot pseudosection¶
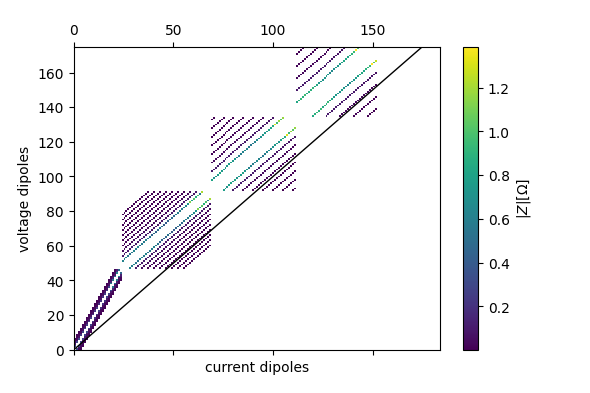
fig = ert.pseudosection('r')
plt.savefig('pseudo_r.png')
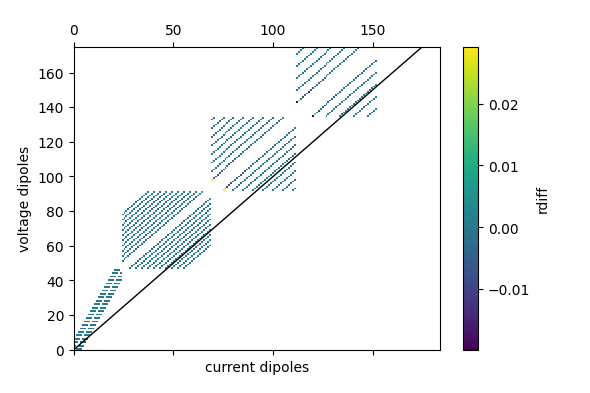
fig = ert.pseudosection('rdiff')
plt.savefig('pseudo_rdiff.png')
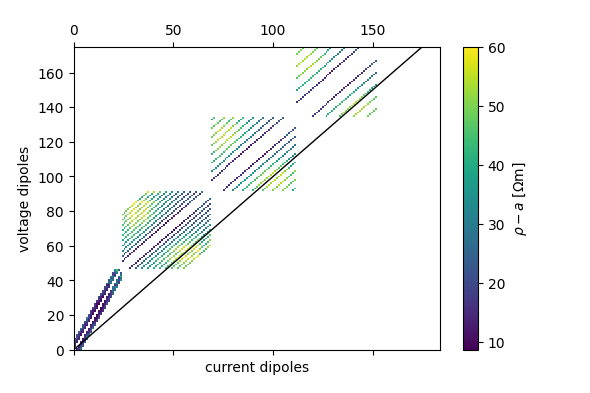
fig = ert.pseudosection('rho_a')
plt.savefig('pseudo_rhoa.png')
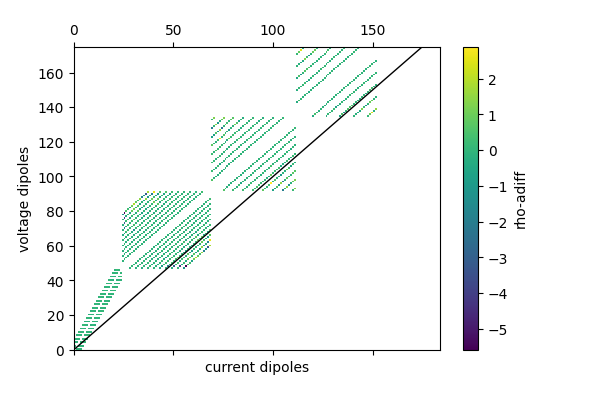
fig = ert.pseudosection('rho_adiff')
plt.savefig('pseudo_rhoadiff.png')
fig = ert.pseudosection('Iab')
plt.savefig('pseudo_I.png')
apply data filters
ert.filter('norrec == "rec"')
ert.filter('r < 0 or r > 1')
ert.filter('rho_a < 0 or rho_a > 60')
ert.filter('rdiff > 0.2 or rdiff < -0.2')
Datenfile in CRTomo Format ausgeben (volt.dat)¶
crto.write_files_to_directory(ert.data, '.')
Arbeiten im Tomodir¶
Gitter und Daten einlesen¶
grid = crtomo.crt_grid.create_surface_grid(
electrodes_x=list(range(30, 174, 3)),
char_lengths=[1.5, 10, 10, 10],
)
td_obj = crtomo.tdMan(grid=grid)
td_obj.read_voltages('volt.dat')
This grid was sorted using CutMcK. The nodes were resorted!
Triangular grid found
Inversionseinstellungen¶
td_obj.crtomo_cfg['robust_inv'] = 'F'
td_obj.crtomo_cfg['dc_inv'] = 'F'
td_obj.crtomo_cfg['cells_z'] = '-1'
td_obj.crtomo_cfg['mag_rel'] = '10'
td_obj.crtomo_cfg['mag_abs'] = '0.5'
td_obj.crtomo_cfg['fpi_inv'] = 'F'
# td_obj.crtomo_cfg['pha_a1'] = '10'
# td_obj.crtomo_cfg['pha_b'] = '-1.5'
# td_obj.crtomo_cfg['pha_rel'] = '10'
# td_obj.crtomo_cfg['pha_abs'] = '0'
invert
td_obj.invert(cores=4)
Attempting inversion in directory: /tmp/tmpb3trxjta
Using binary: /usr/bin/CRTomo_dev
Calling CRTomo
b' ######### CRTomo ############\nLicence:\nCopyright \xc2\xa9 1990-2017 Andreas Kemna <kemna@geo.uni-bonn.de>\nCopyright \xc2\xa9 2008-2017 CRTomo development team (see AUTHORS file)\nPermission is hereby granted, free of charge, to any person obtaining a copy of\nthis software and associated documentation files (the \xe2\x80\x9cSoftware\xe2\x80\x9d), to deal in\nthe Software without restriction, including without limitation the rights to\nuse, copy, modify, merge, publish, distribute, sublicense, and/or sell copies\nof the Software, and to permit persons to whom the Software is furnished to do\nso, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \xe2\x80\x9cAS IS\xe2\x80\x9d, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\nFITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\nAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\nLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\nOUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.\n \n \n Process_ID :: 5892\n OpenMP max threads: 4\n Version control of binary:\n Git-Branch master\n Commit-ID cb89ccd2c4341b9b08bbe7bb08f5aea0d60916ee\n Created Mon-Mar-25-14:34:56-2024\n Compiler \n OS GNU/Linux\n\nOS identification: Linux-OS\n writing inversion results into ../inv\n ## using complex error ellipses ##\n++ check 1\r++ check 2 done!\n + Taking error model\n\n+++ Lambda_0(CRI) = 1940. \n\n Saving lamba presettings -> crt.lamnull\n\n++ (CRI) Setting complex error ellipse\n\n Rescheduling..\n less nodes than wavenumbers\n OpenMP threads: 3( 4)\n\r \r Iteration 0, 0 : Calculating Potentials++ Starting Fit 0.1138 \n\n\n\rWRITING STARTING MODEL\n\n\r \r Iteration 1, 0 : Calculating Sensitivities\nRegularization:: Triangular smooth\n C_m^ calculation time 0d/ 0h/ 0m/ 0s/ 1ms\n\r Iteration 1, 1 : Calculating 1st regularization parameter \r taking easy lam_0 1940. \nlam_0:: 0.19E+04\r \r Iteration 1, 1 : Updating\r \r Iteration 1, 1 : Calculating Potentials -- Update Fit 0.1088 \r \r Iteration 1, 2 : Updating\r \r Iteration 1, 2 : Calculating Potentials -- Update Fit 0.1083 \r \r Iteration 1, 3 : Updating\r \r Iteration 1, 3 : Calculating Potentials -- Update Fit 0.1088 \r \r Iteration 1, 4 : Updating\r \r Iteration 1, 4 : Calculating Potentials -- Update Fit 0.1090 \r \r Iteration 1, 5 : Updating\r \r Iteration 1, 5 : Calculating Potentials -- Update Fit 0.1091 \r \r Iteration 1, 6 : Updating\r \r Iteration 1, 6 : Calculating Potentials -- Update Fit 0.1092 \r \r Iteration 1, 7 : Updating\r \r Iteration 1, 7 : Calculating Potentials -- Update Fit 0.1103 \r \r Iteration 1, 7 : Updating\r \r Iteration 1, 7 : Calculating Potentials ++ Actual Fit 0.1137 \n\n+++ Convergence check (CHI (old/new)) -0.7841E-02 %\n\n\rMODEL ESTIMATE AFTER 1 ITERATIONS \n Iteration terminated: Min. rel. CHI decrease.\n CPU time: 0d/ 0h/ 0m/ 11s/ 139ms\n ***finished***\nNote: The following floating-point exceptions are signalling: IEEE_INVALID_FLAG IEEE_DIVIDE_BY_ZERO\nSTOP 0\n'
Inversion attempt finished
Attempting to import the results
Reading inversion results
is robust False
Info: res_m.diag not found: /tmp/tmpb3trxjta/inv/res_m.diag
/home/runner/work/crtomo_tools/crtomo_tools/examples/06_field_data_processing
Info: ata.diag not found: /tmp/tmpb3trxjta/inv/ata.diag
/home/runner/work/crtomo_tools/crtomo_tools/examples/06_field_data_processing
Statistics of last iteration:
iteration 1
main_iteration 1
it_type DC/IP
type main
dataRMS 0.1137
magRMS 0.1137
phaRMS NaN
lambda 7739.0
roughness 0.0
cgsteps 194.0
nrdata 624.0
steplength 0.001
stepsize 0.00726
l1ratio NaN
Name: 8, dtype: object
save tomodir
td_obj.save_to_tomodir('td')
Total running time of the script: (0 minutes 19.261 seconds)