Note
Go to the end to download the full example code.
Generate Gaussian Models¶
imports
import crtomo
import matplotlib.pylab as plt
we need a tomodir object
grid = crtomo.crt_grid(
'grid_anomalies/elem.dat',
'grid_anomalies/elec.dat',
# 'grid_surface/elem.dat', 'grid_surface/elec.dat'
)
tdm = crtomo.tdMan(grid=grid)
This grid was sorted using CutMcK. The nodes were resorted!
Triangular grid found
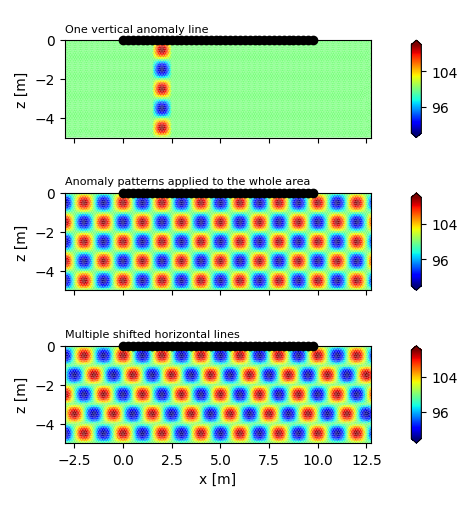
Create a cos(x)cos(y) anomaly
fig, axes = plt.subplots(
3, 1,
sharex=True,
figsize=(12 / 2.54, 13 / 2.54),
)
ax = axes[0]
pid_rmag, rpha = tdm.add_homogeneous_model(100, 0)
p0 = [2.0, -2.5]
anomaly_width = 1
anomaly_height = 1
peak_value = 10
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[2, -0.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
area='only_one_x',
)
tdm.plot.plot_elements_to_ax(
pid_rmag,
ax=ax,
plot_colorbar=True,
cmap_name='jet',
)
ax.set_title('One vertical anomaly line', loc='left', fontsize=8)
ax = axes[1]
pid_rmag, rpha = tdm.add_homogeneous_model(100, 0)
p0 = [2.0, -2.5]
anomaly_width = 1
anomaly_height = 1
peak_value = 10
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[2, -0.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
area='all',
)
tdm.plot.plot_elements_to_ax(
pid_rmag,
ax=ax,
plot_colorbar=True,
cmap_name='jet',
)
ax.set_title(
'Anomaly patterns applied to the whole area', loc='left', fontsize=8)
ax = axes[2]
pid_rmag, rpha = tdm.add_homogeneous_model(100, 0)
p0 = [2.0, -2.5]
anomaly_width = 1
anomaly_height = 1
peak_value = 10
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[0, -0.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
area='only_one_y',
)
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[0.5, -1.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
)
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[1.0, -2.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
)
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[1.5, -3.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
)
tdm.parman.add_2d_cos_anomaly_line(
pid_rmag,
p0=[2.0, -4.5],
anomaly_width=1,
anomaly_height=1,
peak_value=10,
)
# tdm.parman.add_checkerboard_pattern(
# pid_rmag,
# [0, -0.5],
# 1,
# 1,
# 10,
# )
tdm.plot.plot_elements_to_ax(
pid_rmag,
ax=ax,
plot_colorbar=True,
cmap_name='jet',
)
ax.set_title('Multiple shifted horizontal lines', loc='left', fontsize=8)
axes[0].set_xlabel('')
axes[1].set_xlabel('')
fig.tight_layout()
fig.savefig('out_cos.jpg', dpi=300)
# fig, ax = plt.subplots()
# tdm.grid.plot_grid_to_ax(ax)
model already set, will overwrite
model already set, will overwrite
create a new parameter set with one anomaly
create another new parameter set with one anomaly
add an additional anomaly to this parset