Note
Go to the end to download the full example code.
Forward Modeling with CRMod¶
Imports
import crtomo
create a tomodir object from an existing FE mesh
mesh = crtomo.crt_grid(
'grid_surface/g2_large_boundary/elem.dat',
'grid_surface/g2_large_boundary/elec.dat'
)
This grid was sorted using CutMcK. The nodes were resorted!
Triangular grid found
create a tomodir-manager object this object will hold everything required for modeling and inversion runs
tdm = crtomo.tdMan(grid=mesh)
generate measurement configurations
tdm.configs.gen_dipole_dipole(skipc=0)
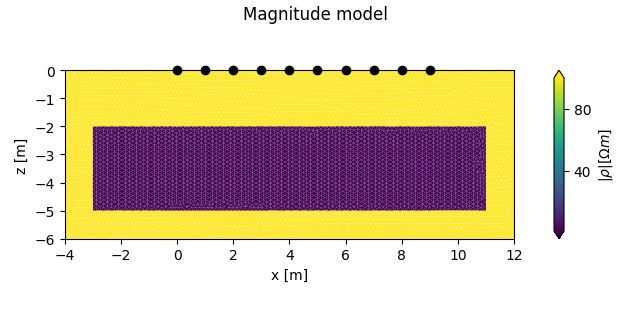
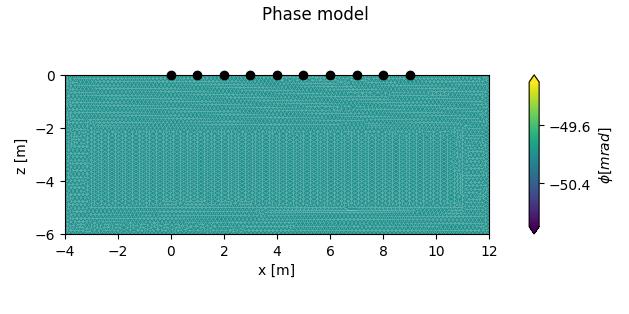
add a forward model with a conductive region
plot the forward models
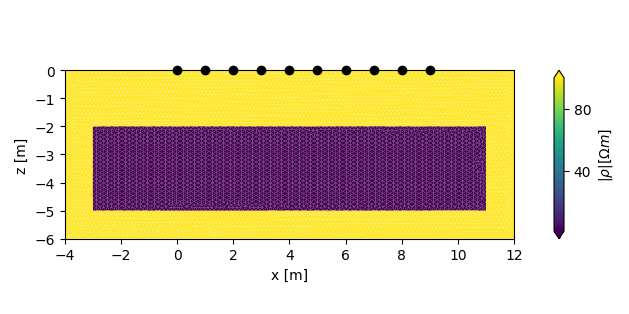
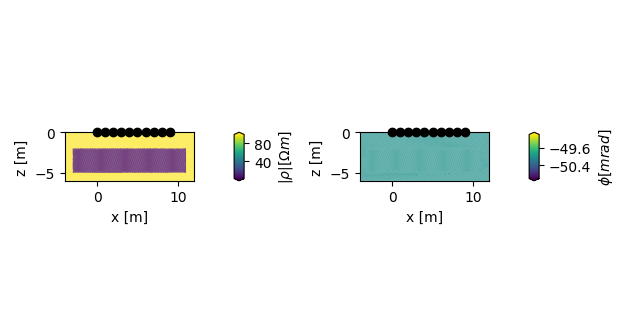
Two lists are returned, each containing either two figures, or two axes
Compute FEM solution using CRMod:
tdm.model(silent=True)
measurements can now be retrieved First column: resistances, second column: phase values [mrad]
measurements = tdm.measurements()
print(measurements)
[[ 5.39044571e+00 -5.00000000e+01]
[ 1.28891766e+00 -5.00000000e+01]
[ 4.36769485e-01 -5.00000000e+01]
[ 1.66427478e-01 -5.00000000e+01]
[ 6.69547841e-02 -5.00000000e+01]
[ 2.78931968e-02 -5.00000000e+01]
[ 1.20029524e-02 -5.00000000e+01]
[ 5.39043760e+00 -5.00000000e+01]
[ 1.28893697e+00 -5.00000000e+01]
[ 4.37060982e-01 -5.00000000e+01]
[ 1.66431203e-01 -5.00000000e+01]
[ 6.69960827e-02 -5.00000000e+01]
[ 2.79575195e-02 -5.00000000e+01]
[ 5.38958788e+00 -5.00000000e+01]
[ 1.28971601e+00 -5.00000000e+01]
[ 4.36941087e-01 -5.00000000e+01]
[ 1.66461006e-01 -5.00000000e+01]
[ 6.70264959e-02 -5.00000000e+01]
[ 5.39728689e+00 -5.00000000e+01]
[ 1.28887296e+00 -5.00000000e+01]
[ 4.36896622e-01 -5.00000000e+01]
[ 1.66363373e-01 -5.00000000e+01]
[ 5.39026976e+00 -5.00000000e+01]
[ 1.28924322e+00 -5.00000000e+01]
[ 4.36632723e-01 -5.00000000e+01]
[ 5.39108038e+00 -5.00000000e+01]
[ 1.28795421e+00 -5.00000000e+01]
[ 5.39027166e+00 -5.00000000e+01]]
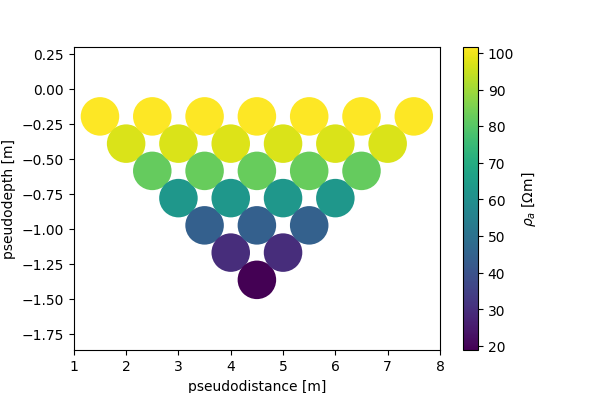
# Let's plot a pseudo-section of the modelled data
ert = tdm.get_fwd_reda_container()
print(ert)
print(ert.data)
# compute geometric factors
ert.compute_K_analytical(spacing=1)
# type 2 pseudosection
fig, ax, cb = ert.pseudosection_type2('rho_a', interpolate=False, spacing=1)
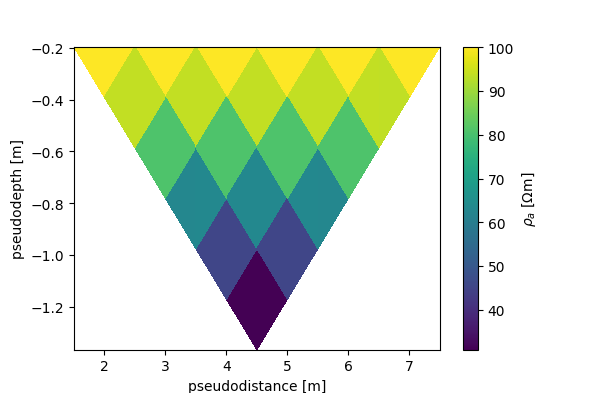
# type 2 pseudosection, interpolated
fig, ax, cb = ert.pseudosection_type2('rho_a', interpolate=True, spacing=1)
# sphinx_gallery_multi_image = "single"
<reda.containers.CR.CR object at 0x7fc2639a6ef0>
a b m n r rpha id norrec rdiff rphadiff
0 1 2 4 3 5.390446 -50.0 0 nor NaN NaN
1 1 2 5 4 1.288918 -50.0 1 nor NaN NaN
2 1 2 6 5 0.436769 -50.0 2 nor NaN NaN
3 1 2 7 6 0.166427 -50.0 3 nor NaN NaN
4 1 2 8 7 0.066955 -50.0 4 nor NaN NaN
5 1 2 9 8 0.027893 -50.0 5 nor NaN NaN
6 1 2 10 9 0.012003 -50.0 6 nor NaN NaN
7 2 3 5 4 5.390438 -50.0 7 nor NaN NaN
8 2 3 6 5 1.288937 -50.0 8 nor NaN NaN
9 2 3 7 6 0.437061 -50.0 9 nor NaN NaN
10 2 3 8 7 0.166431 -50.0 10 nor NaN NaN
11 2 3 9 8 0.066996 -50.0 11 nor NaN NaN
12 2 3 10 9 0.027958 -50.0 12 nor NaN NaN
13 3 4 6 5 5.389588 -50.0 13 nor NaN NaN
14 3 4 7 6 1.289716 -50.0 14 nor NaN NaN
15 3 4 8 7 0.436941 -50.0 15 nor NaN NaN
16 3 4 9 8 0.166461 -50.0 16 nor NaN NaN
17 3 4 10 9 0.067026 -50.0 17 nor NaN NaN
18 4 5 7 6 5.397287 -50.0 18 nor NaN NaN
19 4 5 8 7 1.288873 -50.0 19 nor NaN NaN
20 4 5 9 8 0.436897 -50.0 20 nor NaN NaN
21 4 5 10 9 0.166363 -50.0 21 nor NaN NaN
22 5 6 8 7 5.390270 -50.0 22 nor NaN NaN
23 5 6 9 8 1.289243 -50.0 23 nor NaN NaN
24 5 6 10 9 0.436633 -50.0 24 nor NaN NaN
25 6 7 9 8 5.391080 -50.0 25 nor NaN NaN
26 6 7 10 9 1.287954 -50.0 26 nor NaN NaN
27 7 8 10 9 5.390272 -50.0 27 nor NaN NaN