Note
Go to the end to download the full example code.
Importing FZJ EIT40/EIT160 data¶
This example shows how to import data from the various versions of the EIT system developed by Zimmermann et al. 2008 (http://iopscience.iop.org/article/10.1088/0957-0233/19/9/094010/meta).
At this point we only support 3-point data, i.e., data which uses two electrodes to inject current, and then uses all electrodes to measure the resulting potential distribution against system ground. Classical four-point configurations are then computed using superposition.
Required are two files: a data file (usually eit_data_mnu0.mat and a text file (usually configs.dat containing the measurement configurations to extract.
The configs.dat file contains the four-point spreads to be imported from the measurement. This file is a text file with four columns (A, B, M, N), separated by spaces or tabs. Each line denotes one measurement:
1 2 4 3
2 3 5 6
An alternative to the config.dat file is to permute all current injection dipoles as voltage dipoles, resulting in a fully normal-reciprocal configuration set. This set can be automatically generated from the measurement data by providing a special function as the config-parameter in the import function. This is explained below.
Import reda
import reda
Initialize an sEIT container
seit = reda.sEIT()
# import the data
seit.import_eit_fzj(
filename='data_EIT40_v_EZ-2017/eit_data_mnu0.mat',
configfile='data_EIT40_v_EZ-2017/configs_large_dipoles_norrec.dat'
)
Constructing four-point measurements
Summary:
a b ... frequency rpha
count 16280.0000 16280.0000 ... 16280.000000 16280.000000
mean 10.5250 30.4750 ... 1328.935406 1232.306629
std 5.8096 5.8096 ... 2885.893725 1823.550170
min 1.0000 20.0000 ... 0.100000 -3141.584482
25% 5.7500 25.7500 ... 1.000000 -9.388102
50% 10.5000 30.5000 ... 31.250000 -1.038730
75% 15.2500 35.2500 ... 1000.000000 3132.171261
max 21.0000 40.0000 ... 10000.000000 3141.580651
[8 rows x 7 columns]
an alternative would be to automatically create measurement configurations from the current injection dipoles:
from reda.importers.eit_fzj import MD_ConfigsPermutate
# initialize an sEIT container
seit_not_used = reda.sEIT()
# import the data
seit_not_used.import_eit_fzj(
filename='data_EIT40_v_EZ-2017/eit_data_mnu0.mat',
configfile=MD_ConfigsPermutate
)
Constructing four-point measurements
Summary:
a b ... frequency rpha
count 16280.0000 16280.0000 ... 16280.000000 16280.000000
mean 10.5250 30.4750 ... 1328.935406 217.078193
std 5.8096 5.8096 ... 2885.893725 1129.623431
min 1.0000 20.0000 ... 0.100000 -3141.556931
25% 5.7500 25.7500 ... 1.000000 -15.614618
50% 10.5000 30.5000 ... 31.250000 -7.626176
75% 15.2500 35.2500 ... 1000.000000 -5.227314
max 21.0000 40.0000 ... 10000.000000 3141.584591
[8 rows x 7 columns]
Compute geometric factors
compute normal and reciprocal pairs note that this is usually done on import once.
quadrupoles can be directly accessed using a pandas grouper
print(seit.abmn)
quadpole_data = seit.abmn.get_group((10, 29, 15, 34))
print(quadpole_data[['a', 'b', 'm', 'n', 'frequency', 'r', 'rpha']])
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7fe1af49a320>
a b m n frequency r rpha
748 10 29 15 34 0.100000 7.356375 -5.205543
2228 10 29 15 34 0.314465 7.323810 -6.903037
3708 10 29 15 34 1.000000 7.282164 -7.242394
5188 10 29 15 34 3.125000 7.239073 -6.332998
6668 10 29 15 34 10.000000 7.199660 -12.081013
8148 10 29 15 34 31.250000 7.176920 -3.132819
9628 10 29 15 34 110.000000 7.170031 4.958888
11108 10 29 15 34 312.500000 7.211899 8.671063
12588 10 29 15 34 1000.000000 7.325287 -2.429873
14068 10 29 15 34 3150.000000 7.313380 -37.892727
15548 10 29 15 34 10000.000000 7.035541 -100.904824
in a similar fashion, spectra can be extracted and plotted
spec_nor, spec_rec = seit.get_spectrum(abmn=(10, 29, 15, 34))
print(type(spec_nor))
print(type(spec_rec))
with reda.CreateEnterDirectory('output_eit_fzj'):
spec_nor.plot(filename='spectrum_10_29_15_34.pdf')
# Note that there is also the convenient script
# :py:meth:`reda.sEIT.plot_all_spectra`, which can be used to plot all spectra
# of the container into separate files, given an output directory.
<class 'reda.eis.plots.sip_response'>
<class 'reda.eis.plots.sip_response'>
filter data
seit.remove_frequencies(1e-3, 300)
seit.query('rpha < 10')
seit.query('rpha > -40')
seit.query('rho_a > 15 and rho_a < 35')
seit.query('k < 400')
Remaining frequencies:
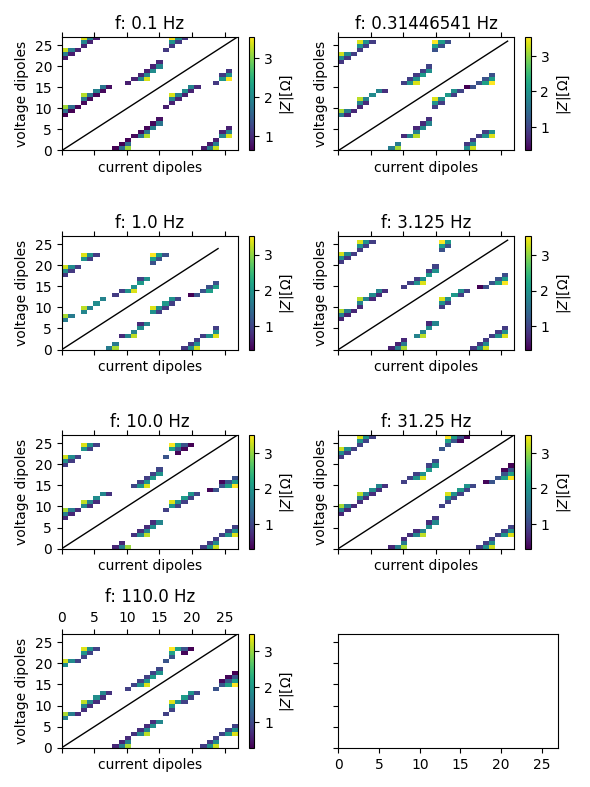
[0.1, 0.31446541, 1.0, 3.125, 10.0, 31.25, 110.0]
Plotting histograms Raw data plots (execute before applying the filters):
# import os
# import reda.plotters.histograms as redahist
# if not os.path.isdir('hists_raw'):
# os.makedirs('hists_raw')
# # plot histograms for all frequencies
# r = redahist.plot_histograms_extra_dims(
# seit.data, ['R', 'rpha'], ['frequency']
# )
# for f in sorted(r.keys()):
# r[f]['all'].savefig('hists_raw/hist_raw_f_{0}.png'.format(f), dpi=300)
# if not os.path.isdir('hists_filtered'):
# os.makedirs('hists_filtered')
# r = redahist.plot_histograms_extra_dims(
# seit.data, ['R', 'rpha'], ['frequency']
# )
# for f in sorted(r.keys()):
# r[f]['all'].savefig(
# 'hists_filtered/hist_filtered_f_{0}.png'.format(f), dpi=300
# )
Now export the data to CRTomo-compatible files this context manager executes all code within the given directory
with reda.CreateEnterDirectory('output_eit_fzj'):
import reda.exporters.crtomo as redaex
redaex.write_files_to_directory(seit.data, 'crt_results', norrec='nor', )
Plot pseudosections of all frequencies
import reda.plotters.pseudoplots as PS
import pylab as plt
with reda.CreateEnterDirectory('output_eit_fzj'):
g = seit.data.groupby('frequency')
fig, axes = plt.subplots(
4, 2,
figsize=(15 / 2.54, 20 / 2.54),
sharex=True, sharey=True
)
for ax, (key, item) in zip(axes.flat, g):
fig, ax, cb = PS.plot_pseudosection_type1(item, ax=ax, column='r')
ax.set_title('f: {} Hz'.format(key))
fig.tight_layout()
fig.savefig('pseudosections_eit40.pdf')
alternative
with reda.CreateEnterDirectory('output_eit_fzj'):
seit.plot_pseudosections(
column='r', filename='pseudosections_eit40_v2.pdf'
)
Total running time of the script: (1 minutes 3.421 seconds)
