Note
Go to the end to download the full example code.
Importing Syscal IP data¶
import reda
a hack to prevent large loading times…
# import os
# import pickle
# pfile = 'ip.pickle'
# if os.path.isfile(pfile):
# with open(pfile, 'rb') as fid:
# ip = pickle.load(fid)
# else:
# ip = reda.TDIP()
# ip.import_syscal_bin('data_syscal_ip/data_normal.bin')
# ip.import_syscal_bin(
# 'data_syscal_ip/data_reciprocal.bin', reciprocals=48)
# with open(pfile, 'wb') as fid:
# pickle.dump(ip, fid)
normal loading of tdip data
ip = reda.TDIP()
# import pprofile
# profiler = pprofile.Profile()
# with profiler():
ip.import_syscal_bin('data_syscal_ip/data_normal.bin')
# profile.print_stats()
# exit()
ip.import_syscal_bin('data_syscal_ip/data_reciprocal.bin', reciprocals=48)
print(ip.data[['a', 'b', 'm', 'n', 'id', 'norrec']])
# import IPython
# IPython.embed()
# exit()
a b m n id norrec
0 1 2 4 5 170 nor
1979 5 4 2 1 170 rec
1978 6 5 2 1 211 rec
1 1 2 5 6 211 nor
44 2 3 5 6 212 nor
... .. .. .. .. ... ...
986 42 43 47 48 1977 nor
988 43 44 47 48 1978 nor
991 48 47 44 43 1978 rec
990 48 47 45 44 1979 rec
989 44 45 47 48 1979 nor
[1980 rows x 6 columns]
plot a decay curve by specifying the index note that no file will be saved to disk if filename parameter is not provided
ip.plot_decay_curve(filename='decay_curve.png',
index_nor=0, index_rec=1978, return_fig=True)
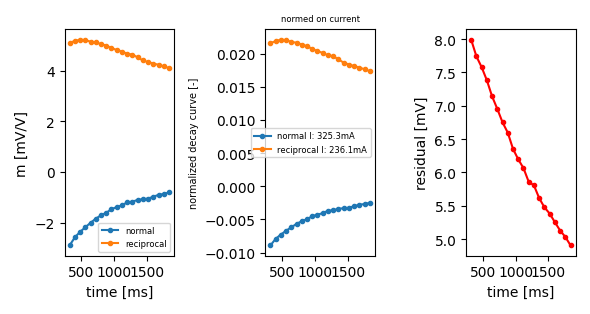
<Figure size 590.551x314.961 with 3 Axes>
you can also specify only one index this will only return a figure object, but will not save to file:
ip.plot_decay_curve(filename='decay_curve.png',
index_nor=0, return_fig=True)
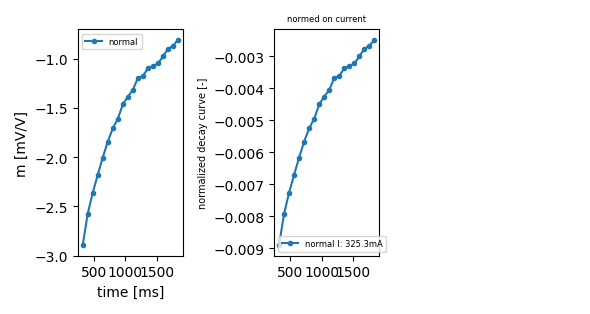
<Figure size 590.551x314.961 with 3 Axes>
it does not matter if you choose normal or reciprocal
ip.plot_decay_curve(index_rec=0, return_fig=True)
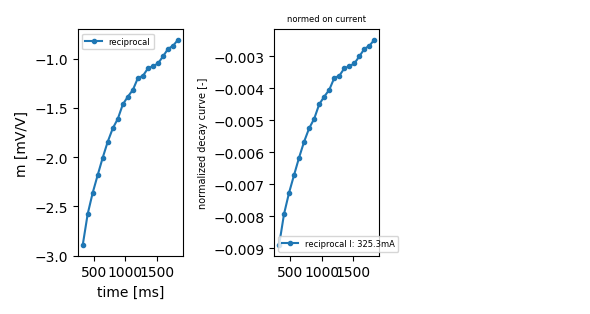
<Figure size 590.551x314.961 with 3 Axes>
plot a decay curve by specifying the index
ip.plot_decay_curve(nr_id=170, return_fig=True)
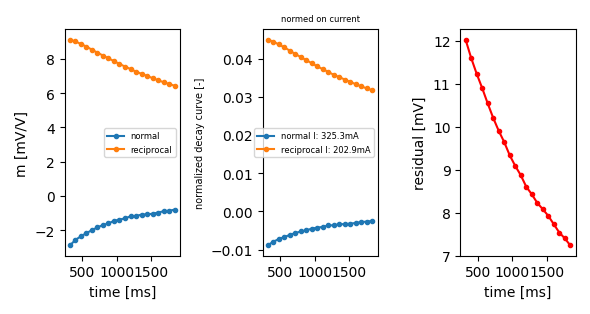
<Figure size 590.551x314.961 with 3 Axes>
a b m n 0 1 2 4 5 1978 5 4 2 1
ip.plot_decay_curve(abmn=(1, 2, 5, 4), return_fig=True)
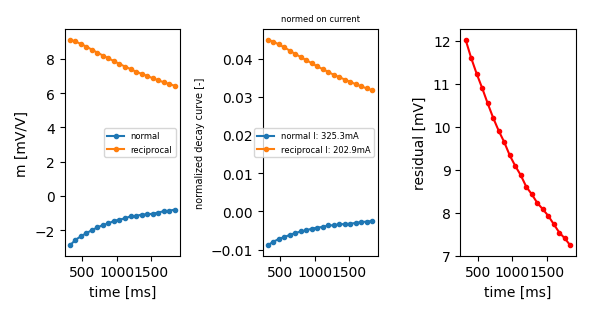
<Figure size 590.551x314.961 with 3 Axes>
reciprocal is also ok
ip.plot_decay_curve(abmn=(4, 5, 2, 1), return_fig=True)
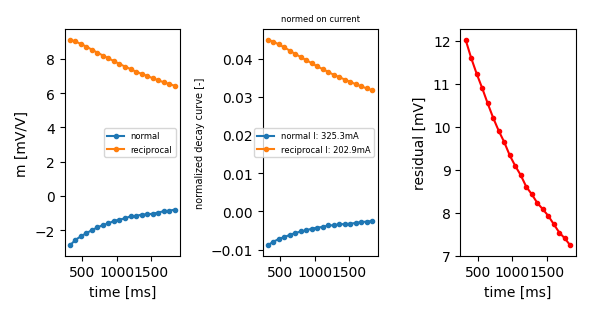
<Figure size 590.551x314.961 with 3 Axes>
