Note
Go to the end to download the full example code.
Importing Syscal ERT data¶
This example is all about import data from IRIS Instruments Syscal systems. There is a variety of different options that should cover most use cases. Also, shortly introduced are the data journal, action log, filtering, and accessing data using the underlying dataframe.
import numpy as np
import matplotlib.pylab as plt
import reda
ert = reda.ERT()
data import:
# note that you should prefer importing the binary data as the text export
# sometimes is missing some of the auxiliary data contained in the binary data.
ert.import_syscal_txt('data_syscal_ert/data_normal.txt')
# the second data set was measured in a reciprocal configuration by switching
# the 24-electrode cables on the Syscal Pro input connectors. The parameter
# "reciprocals" changes electrode notations.
ert.import_syscal_txt(
'data_syscal_ert/data_reciprocal.txt',
reciprocals=48
)
# compute geometrical factors using the analytical half-space equation for a
# spacing of 0.25 m
ert.compute_K_analytical(spacing=0.25)
array([-18.84955592, -18.84955592, -47.1238898 , ..., -47.1238898 ,
-18.84955592, -18.84955592], shape=(1980,))
ert.print_data_journal()
--- Data Journal Start ---
2025-12-09 10:42:38.884880
Data was imported from file (990 data points)
Data was imported from file (990 data points)
--- Data Journal End ---
ert.print_log()
2025-12-09 10:42:37,566 - reda.containers.BaseContainer - INFO - IRIS Syscal Pro text import
2025-12-09 10:42:38,169 - reda.containers.BaseContainer - INFO - Data sized changed from 0 to 990
2025-12-09 10:42:38,169 - reda.containers.BaseContainer - INFO - IRIS Syscal Pro text import
2025-12-09 10:42:38,878 - reda.containers.BaseContainer - INFO - Data sized changed from 990 to 1980
create some plots in a subdirectory
with reda.CreateEnterDirectory('plots'):
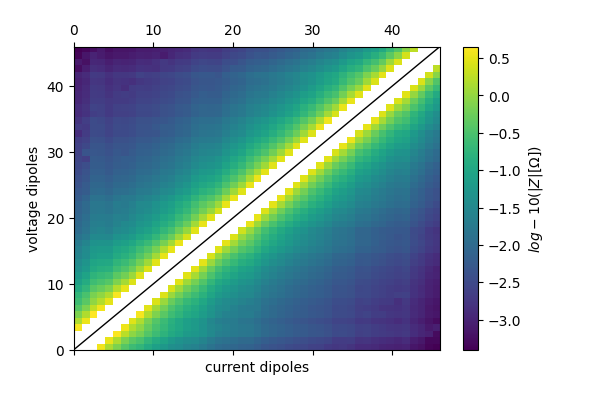
ert.pseudosection(
column='r', filename='pseudosection_log10_r.pdf', log10=True)
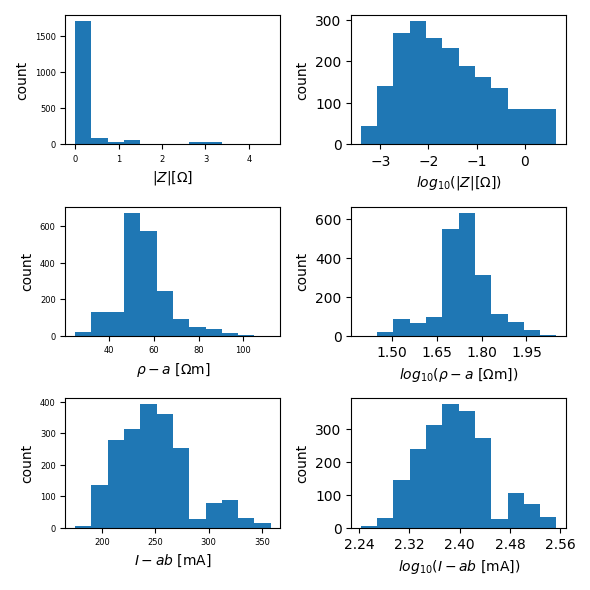
ert.histogram(['r', 'rho_a', 'Iab', ], filename='histograms.pdf')
export to various data files
with reda.CreateEnterDirectory('output_01_syscal_import'):
ert.export_bert('data.ohm')
ert.export_pygimli('data.pygimli')
ert.export_crtomo('volt.dat')
The data is internally stored in a pandas.DataFrame As such, you can always use the data directly and build your custom functionality on top of REDA
print(ert.data)
a b m n Iab ... norrec rdiff k rho_a sigma_a
0 1 2 5 4 325.250 ... nor -0.248598 18.849556 73.639666 0.013580
1979 4 5 2 1 202.874 ... rec -0.248598 18.849556 68.953711 0.014502
1978 5 6 2 1 236.119 ... rec -0.065825 47.123890 63.731679 0.015691
1 1 2 6 5 325.250 ... nor -0.065825 47.123890 66.833629 0.014963
44 2 3 6 5 256.492 ... nor -0.144387 18.849556 63.872224 0.015656
... .. .. .. .. ... ... ... ... ... ... ...
986 42 43 48 47 174.871 ... nor -0.000643 94.247780 33.877159 0.029518
988 43 44 48 47 179.834 ... nor -0.006144 47.123890 39.976982 0.025014
991 47 48 44 43 245.897 ... rec -0.006144 47.123890 39.687461 0.025197
990 47 48 45 44 245.897 ... rec -0.049032 18.849556 32.889082 0.030405
989 44 45 48 47 242.947 ... nor -0.049032 18.849556 33.813317 0.029574
[1980 rows x 13 columns]
Lets apply an arbitrary filter. Note that the change in data numbers is logged. You can use all columns defined in the data frame for more complex filters
ert.filter('r <= 0')
ert.filter('(a == 1) and Iab <= 100')
ert.print_data_journal()
--- Data Journal Start ---
2025-12-09 10:42:41.460651
Data was imported from file (990 data points)
Data was imported from file (990 data points)
A filter was applied with query "r <= 0". In total 0 records were removed
A filter was applied with query "(a == 1) and Iab <= 100". In total 0 records were removed
--- Data Journal End ---
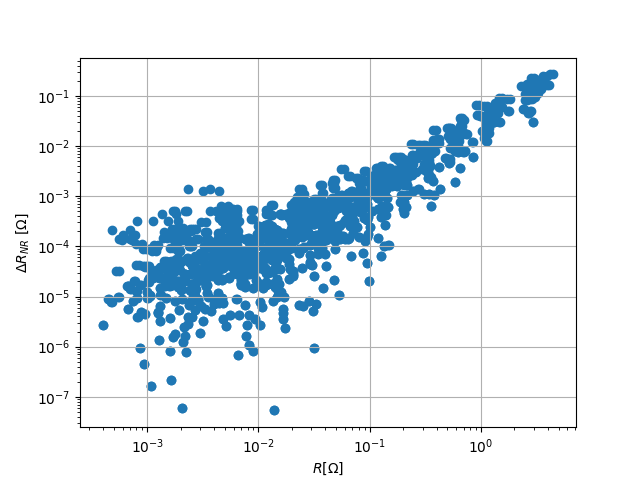
Also note that normal-reciprocal differences were directly computed.
fig, ax = plt.subplots()
ax.scatter(
ert.data['r'],
np.abs(ert.data['rdiff']),
)
ax.set_xlabel(r'$R [\Omega$]')
ax.set_ylabel(r'$\Delta R_{NR}~[\Omega$]')
ax.grid()
ax.set_xscale('log')
ax.set_yscale('log')
The column ‘id’ groups quadrupoles belonging to the same normal-reciprocal pair. For example, plot some of the groups
Id: 129
a b m n r rho_a k Iab
0 1 2 5 4 3.906706 73.639666 18.849556 325.250
1 4 5 2 1 3.658108 68.953711 18.849556 202.874
Id: 171
a b m n r rho_a k Iab
2 5 6 2 1 1.352428 63.731679 47.12389 236.119
3 1 2 6 5 1.418254 66.833629 47.12389 325.250
Id: 172
a b m n r rho_a k Iab
5 2 3 6 5 3.388527 63.872224 18.849556 256.492
4 5 6 3 2 3.244140 61.150591 18.849556 236.119
Id: 213
a b m n r rho_a k Iab
7 1 2 7 6 0.725030 68.332465 94.24778 325.250
6 6 7 2 1 0.717153 67.590078 94.24778 207.264
Id: 214
a b m n r rho_a k Iab
8 6 7 3 2 1.442918 67.995919 47.12389 207.264
9 2 3 7 6 1.473301 69.427689 47.12389 256.492
Id: 215
a b m n r rho_a k Iab
10 3 4 7 6 4.500319 84.829014 18.849556 239.825
11 6 7 4 3 4.233244 79.794762 18.849556 207.264
There are various ways to import Syscal data, relating to the electrode numbering:
ert1 = reda.ERT()
ert1.import_syscal_bin(
'data_syscal_ert/02_data_normal_thinned_not_all_electrodes/data.bin',
check_meas_nums=False,
)
print(ert1.electrode_positions)
ert2 = reda.ERT()
ert2.import_syscal_bin(
'data_syscal_ert/02_data_normal_thinned_not_all_electrodes/data.bin',
check_meas_nums=False,
elecs_transform_reg_spacing_x=(1, 2.5),
)
print(ert2.electrode_positions)
ert3 = reda.ERT()
ert3.import_syscal_bin(
'data_syscal_ert/02_data_normal_thinned_not_all_electrodes/data.bin',
check_meas_nums=False,
assume_regular_electrodes_x=(48, 1.0),
# elecs_transform_reg_spacing_x=(1, 2.5),
)
print(ert3.electrode_positions)
ert_rec = reda.ERT()
ert_rec.import_syscal_bin(
'data_syscal_ert/02_data_normal_thinned_not_all_electrodes/data.bin',
check_meas_nums=False,
assume_regular_electrodes_x=(48, 1.0),
elecs_transform_reg_spacing_x=(1, 2.5),
reciprocals=48,
)
print(ert_rec.electrode_positions)
x y z
electrode_number
1 0.0 0.0 0.0
2 1.0 0.0 0.0
3 3.0 0.0 0.0
4 4.0 0.0 0.0
5 5.0 0.0 0.0
6 6.0 0.0 0.0
7 7.0 0.0 0.0
8 8.0 0.0 0.0
9 9.0 0.0 0.0
10 10.0 0.0 0.0
11 11.0 0.0 0.0
12 12.0 0.0 0.0
13 40.0 0.0 0.0
14 41.0 0.0 0.0
15 42.0 0.0 0.0
16 43.0 0.0 0.0
17 44.0 0.0 0.0
18 45.0 0.0 0.0
19 46.0 0.0 0.0
20 47.0 0.0 0.0
x y z
electrode_number
1 0.0 0.0 0.0
2 2.5 0.0 0.0
3 7.5 0.0 0.0
4 10.0 0.0 0.0
5 12.5 0.0 0.0
6 15.0 0.0 0.0
7 17.5 0.0 0.0
8 20.0 0.0 0.0
9 22.5 0.0 0.0
10 25.0 0.0 0.0
11 27.5 0.0 0.0
12 30.0 0.0 0.0
13 100.0 0.0 0.0
14 102.5 0.0 0.0
15 105.0 0.0 0.0
16 107.5 0.0 0.0
17 110.0 0.0 0.0
18 112.5 0.0 0.0
19 115.0 0.0 0.0
20 117.5 0.0 0.0
x y z
electrode_number
1 0.0 0.0 0.0
2 1.0 0.0 0.0
3 2.0 0.0 0.0
4 3.0 0.0 0.0
5 4.0 0.0 0.0
6 5.0 0.0 0.0
7 6.0 0.0 0.0
8 7.0 0.0 0.0
9 8.0 0.0 0.0
10 9.0 0.0 0.0
11 10.0 0.0 0.0
12 11.0 0.0 0.0
13 12.0 0.0 0.0
14 13.0 0.0 0.0
15 14.0 0.0 0.0
16 15.0 0.0 0.0
17 16.0 0.0 0.0
18 17.0 0.0 0.0
19 18.0 0.0 0.0
20 19.0 0.0 0.0
21 20.0 0.0 0.0
22 21.0 0.0 0.0
23 22.0 0.0 0.0
24 23.0 0.0 0.0
25 24.0 0.0 0.0
26 25.0 0.0 0.0
27 26.0 0.0 0.0
28 27.0 0.0 0.0
29 28.0 0.0 0.0
30 29.0 0.0 0.0
31 30.0 0.0 0.0
32 31.0 0.0 0.0
33 32.0 0.0 0.0
34 33.0 0.0 0.0
35 34.0 0.0 0.0
36 35.0 0.0 0.0
37 36.0 0.0 0.0
38 37.0 0.0 0.0
39 38.0 0.0 0.0
40 39.0 0.0 0.0
41 40.0 0.0 0.0
42 41.0 0.0 0.0
43 42.0 0.0 0.0
44 43.0 0.0 0.0
45 44.0 0.0 0.0
46 45.0 0.0 0.0
47 46.0 0.0 0.0
48 47.0 0.0 0.0
x y z
electrode_number
1 0.0 0.0 0.0
2 2.5 0.0 0.0
3 5.0 0.0 0.0
4 7.5 0.0 0.0
5 10.0 0.0 0.0
6 12.5 0.0 0.0
7 15.0 0.0 0.0
8 17.5 0.0 0.0
9 20.0 0.0 0.0
10 22.5 0.0 0.0
11 25.0 0.0 0.0
12 27.5 0.0 0.0
13 30.0 0.0 0.0
14 32.5 0.0 0.0
15 35.0 0.0 0.0
16 37.5 0.0 0.0
17 40.0 0.0 0.0
18 42.5 0.0 0.0
19 45.0 0.0 0.0
20 47.5 0.0 0.0
21 50.0 0.0 0.0
22 52.5 0.0 0.0
23 55.0 0.0 0.0
24 57.5 0.0 0.0
25 60.0 0.0 0.0
26 62.5 0.0 0.0
27 65.0 0.0 0.0
28 67.5 0.0 0.0
29 70.0 0.0 0.0
30 72.5 0.0 0.0
31 75.0 0.0 0.0
32 77.5 0.0 0.0
33 80.0 0.0 0.0
34 82.5 0.0 0.0
35 85.0 0.0 0.0
36 87.5 0.0 0.0
37 90.0 0.0 0.0
38 92.5 0.0 0.0
39 95.0 0.0 0.0
40 97.5 0.0 0.0
41 100.0 0.0 0.0
42 102.5 0.0 0.0
43 105.0 0.0 0.0
44 107.5 0.0 0.0
45 110.0 0.0 0.0
46 112.5 0.0 0.0
47 115.0 0.0 0.0
48 117.5 0.0 0.0