Note
Go to the end to download the full example code.
Exporting to SimPEG for ERT inversion¶
Example that shows how to export data to SimPEG
- This example basically reuses, and simplifies, the following SimPEG tutorial:
https://simpeg.xyz/user-tutorials/inv-dcr-2d#define-and-run-the-inversion
Work in progress example!
sphinx_gallery_thumbnail_number = 4
import reda
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
simpeg-specific imports
from simpeg.data import Data
from simpeg.electromagnetics.static.utils.static_utils import (
plot_pseudosection,
)
from discretize import TreeMesh
from discretize.utils import active_from_xyz
from simpeg.electromagnetics.static.utils import (
generate_survey_from_abmn_locations,
)
from simpeg import (
maps,
# data,
data_misfit,
regularization,
optimization,
inverse_problem,
inversion,
directives,
)
from simpeg.electromagnetics.static import resistivity as dc
mpl.use('Agg')
# import warnings
# import simpeg
# warnings.filterwarnings(
# 'ignore', simpeg.utils.solver_utils.DefaultSolverWarning
# )
import data into reda, including electrode information
data_e = reda.ERT()
data_e.import_syscal_bin('../01_ERT/data_rodderberg/20140208_01.bin')
data_e.import_electrode_positions(
'../01_ERT/data_rodderberg/electrode_positions.dat',
)
# plot the electrode positions
# data.plot_electrode_positions_2d()
- with reda.CreateEnterDirectory(‘output_01_ertinv’):
data.histogram(‘r’, log10=True, filename=’histograms_raw.pdf’)
data_e.compute_K_numerical(
{
'rho': 100,
'elem': '../01_ERT/data_rodderberg/mesh_creation/g1/elem.dat',
'elec': '../01_ERT/data_rodderberg/mesh_creation/g1/elec.dat',
}
)
data_e.filter('rho_a <= 0')
apparent_resistivities = data_e.data['rho_a'].values
# normalized voltages, also called transfer resistances
data_volt = data_e.data['r'].values
SETTINGS
{'rho': 100, 'elem': '../01_ERT/data_rodderberg/mesh_creation/g1/elem.dat', 'elec': '../01_ERT/data_rodderberg/mesh_creation/g1/elec.dat'}
a_locs = data_e.electrode_positions.iloc[data_e.data['a'] - 1, [0, 2]].values
b_locs = data_e.electrode_positions.iloc[data_e.data['b'] - 1, [0, 2]].values
m_locs = data_e.electrode_positions.iloc[data_e.data['m'] - 1, [0, 2]].values
n_locs = data_e.electrode_positions.iloc[data_e.data['n'] - 1, [0, 2]].values
survey, out_indices = generate_survey_from_abmn_locations(
locations_a=a_locs,
locations_b=b_locs,
locations_m=m_locs,
locations_n=n_locs,
data_type="volt",
output_sorting=True,
)
data_object = Data(survey, dobs=data_volt)
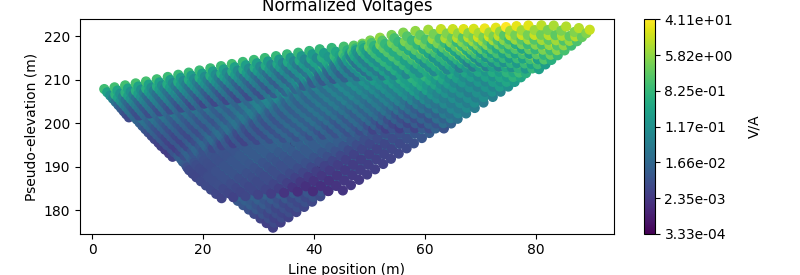
# Plot voltages pseudo-section
fig = plt.figure(figsize=(8, 2.75))
ax1 = fig.add_axes([0.1, 0.15, 0.75, 0.78])
plot_pseudosection(
data_object,
plot_type="scatter",
ax=ax1,
scale="log",
cbar_label="V/A",
scatter_opts={"cmap": mpl.cm.viridis},
)
ax1.set_title("Normalized Voltages")
with reda.CreateEnterDirectory('output_00_ertinv'):
fig.savefig('pseudosection.jpg', dpi=300)
data_object.standard_deviation = 7e-5 + 0.01 * np.abs(data_object.dobs)
https://simpeg.xyz/user-tutorials/inv-dcr-2d#define-and-run-the-inversion
dh = 1 # base cell width
dom_width_x = 150.0 # domain width x
dom_width_z = 40.0 # domain width z
nbcx = 2 ** int(
np.round(np.log(dom_width_x / dh) / np.log(2.0))) # num. base cells x
nbcz = 2 ** int(
np.round(np.log(dom_width_z / dh) / np.log(2.0))) # num. base cells z
# Define the base mesh with top at z = 0 m
hx = [(dh, nbcx)]
hz = [(dh, nbcz)]
mesh = TreeMesh(
[hx, hz],
origin=('0', '0'),
diagonal_balance=True,
)
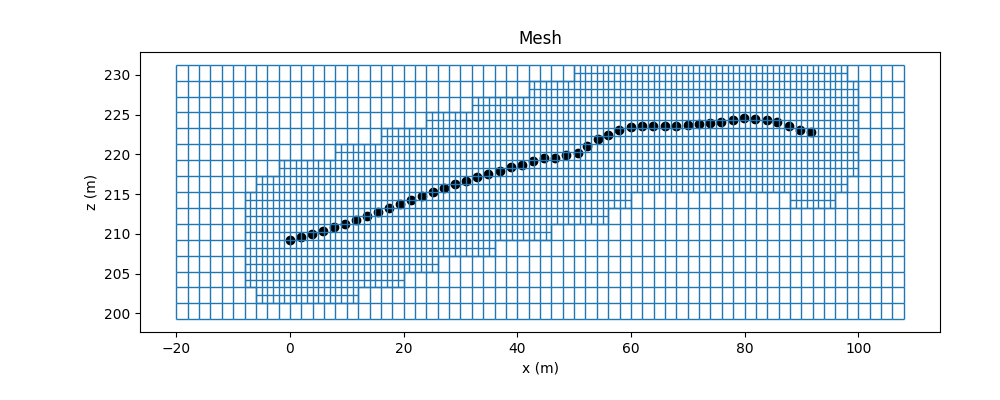
topo_2d = data_e.electrode_positions[['x', 'z']].values
x_dist = np.abs(topo_2d[:, 0].max() - topo_2d[:, 0].min())
# Shift top to maximum topography
mesh.origin = mesh.origin + np.r_[-20, topo_2d[:, 1].min() - 10]
# Mesh refinement based on topography
mesh.refine_surface(
topo_2d,
padding_cells_by_level=[0, 0, 4, 4],
finalize=False,
)
# Extract unique electrode locations.
unique_locations = data_object.survey.unique_electrode_locations
# Mesh refinement near electrodes.
mesh.refine_points(
unique_locations,
padding_cells_by_level=[8, 12, 6, 6],
finalize=False,
)
mesh.finalize()
fig = plt.figure(figsize=(10, 4))
ax1 = fig.add_axes([0.14, 0.17, 0.8, 0.7])
mesh.plot_grid(ax=ax1, linewidth=1)
ax1.grid(False)
# ax1.set_xlim(-1500, 1500)
# ax1.set_ylim(np.max(z_topo) - 1000, np.max(z_topo))
ax1.set_title("Mesh")
ax1.set_xlabel("x (m)")
ax1.set_ylabel("z (m)")
ax1.scatter(topo_2d[:, 0], topo_2d[:, 1], color='k')
plt.show()
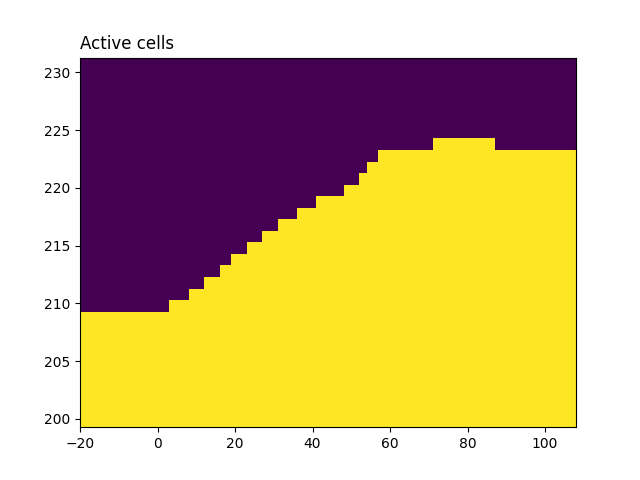
Indices of the active mesh cells from topography (e.g. cells below surface)
active_cells = active_from_xyz(mesh, topo_2d)
# number of active cells
n_active = np.sum(active_cells)
survey.drape_electrodes_on_topography(mesh, active_cells, option="top")
fig, ax = plt.subplots()
mesh.plot_image(active_cells, ax=ax)
ax.set_title('Active cells', loc='left')
fig.show()
Map model parameters to all cells
log_conductivity_map = maps.InjectActiveCells(
mesh, active_cells, 1e-8) * maps.ExpMap(
nP=n_active
)
# Median apparent resistivity
median_resistivity = np.median(apparent_resistivities)
# Create starting model from log-conductivity
starting_conductivity_model = np.log(
1 / median_resistivity) * np.ones(n_active)
# Zero reference conductivity model
reference_conductivity_model = starting_conductivity_model.copy()
voltage_simulation = dc.simulation_2d.Simulation2DNodal(
mesh,
survey=data_object.survey,
sigmaMap=log_conductivity_map,
storeJ=True,
)
dmis_L2 = data_misfit.L2DataMisfit(
simulation=voltage_simulation,
data=data_object
)
reg_L2 = regularization.WeightedLeastSquares(
mesh,
active_cells=active_cells,
alpha_s=dh**-2,
alpha_x=1,
alpha_y=1,
reference_model=reference_conductivity_model,
reference_model_in_smooth=False,
)
# WARNING: maximum number of iterations set to 2!!!!
opt_L2 = optimization.InexactGaussNewton(
maxIter=2,
maxIterLS=20,
maxIterCG=20,
tolCG=1e-3,
)
inv_prob_L2 = inverse_problem.BaseInvProblem(dmis_L2, reg_L2, opt_L2)
sensitivity_weights = directives.UpdateSensitivityWeights(
every_iteration=True,
threshold_value=1e-2
)
update_jacobi = directives.UpdatePreconditioner(update_every_iteration=True)
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=10)
beta_schedule = directives.BetaSchedule(coolingFactor=2.0, coolingRate=2)
target_misfit = directives.TargetMisfit(chifact=1.0)
directives_list_L2 = [
sensitivity_weights,
update_jacobi,
starting_beta,
beta_schedule,
target_misfit,
]
# Here we combine the inverse problem and the set of directives
inv_L2 = inversion.BaseInversion(inv_prob_L2, directives_list_L2)
# Run the inversion
# recovered_model_L2 = inv_L2.run(np.log(0.01) * np.ones(n_param))
recovered_log_conductivity_model = inv_L2.run(starting_conductivity_model)
Running inversion with SimPEG v0.24.0
/home/runner/.virtualenvs/reda/lib/python3.10/site-packages/simpeg/base/pde_simulation.py:490: DefaultSolverWarning: Using the default solver: SolverLU.
If you would like to suppress this notification, add
warnings.filterwarnings('ignore', simpeg.utils.solver_utils.DefaultSolverWarning)
to your script.
return get_default_solver(warn=True)
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using same Solver, and solver_opts as the Simulation2DNodal problem***
/home/runner/.virtualenvs/reda/lib/python3.10/site-packages/pymatsolver/wrappers.py:79: UnusedArgumentWarning: Unused keyword argument "is_symmetric" for splu.
self.kwargs = kwargs
/home/runner/.virtualenvs/reda/lib/python3.10/site-packages/pymatsolver/wrappers.py:79: UnusedArgumentWarning: Unused keyword argument "is_positive_definite" for splu.
self.kwargs = kwargs
/home/runner/.virtualenvs/reda/lib/python3.10/site-packages/pymatsolver/wrappers.py:81: SparseEfficiencyWarning: splu converted its input to CSC format
self.solver = fun(self.A, **self.kwargs)
/home/runner/.virtualenvs/reda/lib/python3.10/site-packages/pymatsolver/solvers.py:415: FutureWarning: In Future pymatsolver v0.4.0, passing a vector of shape (n, 1) to the solve method will return an array with shape (n, 1), instead of always returning a flattened array. This is to be consistent with numpy.linalg.solve broadcasting.
return self.solve(val)
model has any nan: 0
============================ Inexact Gauss Newton ============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 2.56e+06 2.87e+07 0.00e+00 2.87e+07 4.27e+06 0
1 2.56e+06 1.95e+07 1.32e+00 2.29e+07 3.20e+06 0
2 1.28e+06 1.53e+07 2.07e+00 1.80e+07 9.21e+05 0 Skip BFGS
------------------------- STOP! -------------------------
0 : |fc-fOld| = 4.9594e+06 <= tolF*(1+|f0|) = 2.8736e+06
1 : |xc-x_last| = 3.7124e+00 <= tolX*(1+|x0|) = 1.6704e+01
0 : |proj(x-g)-x| = 9.2056e+05 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.2056e+05 <= 1e3*eps = 1.0000e-02
1 : maxIter = 2 <= iter = 2
------------------------- DONE! -------------------------
Plot the model
recovered_conductivity_L2 = np.exp(recovered_log_conductivity_model)
rec_res = 1 / recovered_conductivity_L2
plotting_map = maps.InjectActiveCells(mesh, active_cells, np.nan)
fig = plt.figure(figsize=(9, 4))
# norm = mpl.colors.Normalize(vmin=20, vmax=100)
norm = mpl.colors.LogNorm(vmin=1e0, vmax=1e3)
ax1 = fig.add_axes([0.14, 0.17, 0.68, 0.7])
mesh.plot_image(
plotting_map * rec_res,
ax=ax1,
grid=False,
# clim=[20, 100],
pcolor_opts={
"norm": norm,
"cmap": mpl.cm.RdYlBu_r
},
)
ax1.set_title('SimPEG inversion result', loc='left')
ax2 = fig.add_axes([0.84, 0.17, 0.03, 0.7])
cbar = mpl.colorbar.ColorbarBase(
ax2,
norm=norm,
orientation="vertical",
cmap=mpl.cm.RdYlBu_r,
)
with reda.CreateEnterDirectory('output_00_ertinv'):
fig.savefig('simpeg_inversion_result.jpg', dpi=300)
print('Min/max resistivities:', rec_res.min(), rec_res.max())
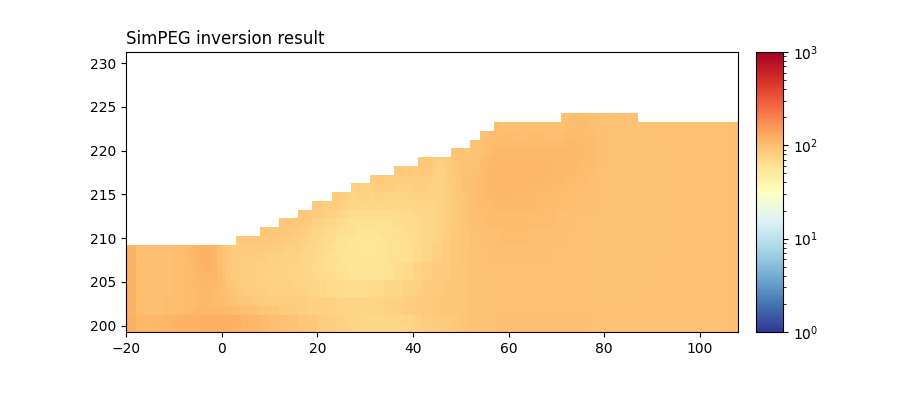
Min/max resistivities: 56.42575696453461 123.98623874271505
Total running time of the script: (1 minutes 28.423 seconds)
