Note
Go to the end to download the full example code.
Exporting to pygimli for ERT inversion¶
REDA can also export to pygimli (https://www.pygimli.org/) ert data containers.
# sphinx_gallery_thumbnail_number = 3
import reda
import pygimli as pg
from pygimli.physics import ert
import data into reda, including electrode information
data = reda.ERT()
data.import_syscal_bin('../01_ERT/data_rodderberg/20140208_01.bin')
data.import_electrode_positions(
'../01_ERT/data_rodderberg/electrode_positions.dat',
)
# plot the electrode positions
data.plot_electrode_positions_2d()
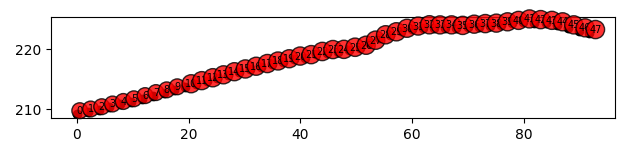
09/12/25 - 10:46:25 - reda.containers.BaseContainer - INFO - IRIS Syscal Pro bin import
09/12/25 - 10:46:27 - reda.containers.BaseContainer - INFO - Data sized changed from 0 to 2816
(<Figure size 629.921x157.48 with 1 Axes>, None)
compute geometric factors
data.compute_K_numerical(fem_code='pygimli')
COMPUTING GEomFacs using PYGimli
Computing numerical geometric factors using Pyglimi
09/12/25 - 10:46:27 - pyGIMLi - INFO - Create default mesh for geometric factor calculation.
09/12/25 - 10:46:28 - pyGIMLi - INFO - Save RVector binary
09/12/25 - 10:46:28 - pyGIMLi - INFO - Cache stored: /home/runner/.cache/pygimli/2165456023755851530
array([ -38.52402364, -153.52866075, -378.10597767, ..., -38.54919138,
-171.86663742, -42.25692476], shape=(2816,))
apply some light filtering
data.filter('rho_a <= 0')
09/12/25 - 10:46:28 - reda.containers.BaseContainer - INFO - Data sized changed from 2816 to 2816
<reda.containers.ERT.ERT object at 0x7fe1d28d74c0>
plot histograms
with reda.CreateEnterDirectory('output_01_ertinv'):
data.histogram('r', log10=True, filename='histograms_raw.jpg')
export the data (and electrode positions) into a pygimli data container
pg_scheme = data.export_to_pygimli_scheme()
WARNING: Implementation and testing still in progress!!!!
WARNING: Journal and log is not copied!
set data errors
pg_scheme.estimateError(relativeError=0.01, absoluteUError=7e-5)
mgr = ert.ERTManager(pg_scheme)
with reda.CreateEnterDirectory('output_01_ertinv'):
mod = mgr.invert(
pg_scheme,
lam=10,
verbose=True,
paraDX=0.1,
paraMaxCellSize=10,
paraDepth=20,
quality=33.0
)
09/12/25 - 10:46:30 - pyGIMLi - INFO - Found 2 regions.
09/12/25 - 10:46:30 - pyGIMLi - INFO - Region with smallest marker set to background (marker=1)
09/12/25 - 10:46:30 - pyGIMLi - INFO - Found 2 regions.
09/12/25 - 10:46:30 - pyGIMLi - INFO - Region with smallest marker set to background (marker=1)
09/12/25 - 10:46:30 - pyGIMLi - INFO - Creating forward mesh from region infos.
09/12/25 - 10:46:30 - pyGIMLi - INFO - Creating refined mesh (H2) to solve forward task.
09/12/25 - 10:46:31 - pyGIMLi - INFO - Mesh for forward task: Mesh: Nodes: 5239 Cells: 9904 Boundaries: 7714
09/12/25 - 10:46:31 - pyGIMLi - INFO - Use median(data values)=96.18936845699801
09/12/25 - 10:46:31 - pyGIMLi - INFO - Created startmodel from forward operator:2008, min/max=96.189368/96.189368
09/12/25 - 10:46:31 - pyGIMLi - INFO - Starting inversion.
fop: <pygimli.physics.ert.ertModelling.ERTModelling object at 0x7fe1a11b4220>
Data transformation: Logarithmic LU transform, lower bound 0.0, upper bound 0.0
Model transformation: Logarithmic transform
min/max (data): 1.86/2230
min/max (error): 1%/211%
min/max (start model): 96.19/96.19
--------------------------------------------------------------------------------
inv.iter 0 ... chi² = 6933.37
--------------------------------------------------------------------------------
inv.iter 1 ... chi² = 138.94 (dPhi = 97.99%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 2 ... chi² = 18.34 (dPhi = 86.70%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 3 ... chi² = 1.65 (dPhi = 89.64%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 4 ... chi² = 1.44 (dPhi = 11.72%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 5 ... chi² = 1.22 (dPhi = 13.79%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 6 ... chi² = 1.06 (dPhi = 10.36%) lam: 10.0
--------------------------------------------------------------------------------
inv.iter 7 ... chi² = 1.06 (dPhi = 0.09%) lam: 10.0
################################################################################
# Abort criterion reached: dPhi = 0.09 (< 2.0%) #
################################################################################
plot the result
with reda.CreateEnterDirectory('output_01_ertinv'):
ax, cb = mgr.showResult(
cMap="Spectral_r",
logScale=False,
)
ax.set_title('PyGimli inversion result', loc='left')
fig = ax.get_figure()
fig.savefig('pygimli_inversion.jpg', dpi=300)
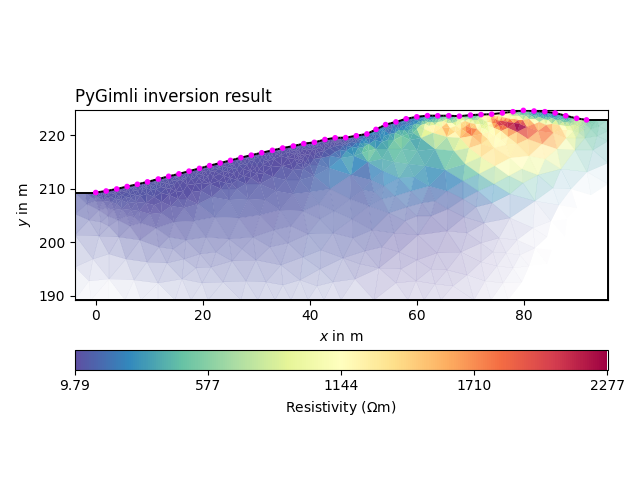
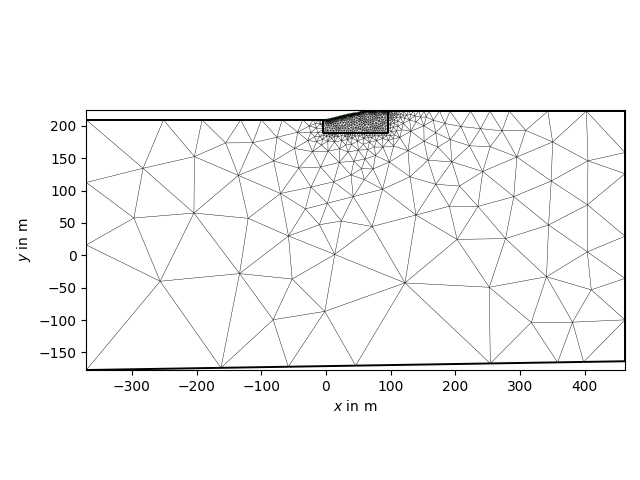
with reda.CreateEnterDirectory('output_01_ertinv'):
ax, _ = pg.show(mgr.mesh)
ax.get_figure().savefig('mesh.jpg', dpi=1200)
Total running time of the script: (2 minutes 14.374 seconds)
